The Tidymodels Approach: Recipes and Workflows
How data science is done in R has changed rapidly over the last few years with the introduction of the tidyverse and tidymodelling.
Many concepts like recipes and workflows are quite abstract and difficult to grasp initially, especially if you are new to data science and coding. That’s totally normal.
However, once you truly understand them, you will realise how they can save you from writing messy and difficult-to-follow code.
In this blog post, we want to address the confusion around recipes and workflows that probably still exist.
The following visual provides a minimal solution with all the steps we have gone through in this course so far. See below for a step-by-step breakdown of this process.
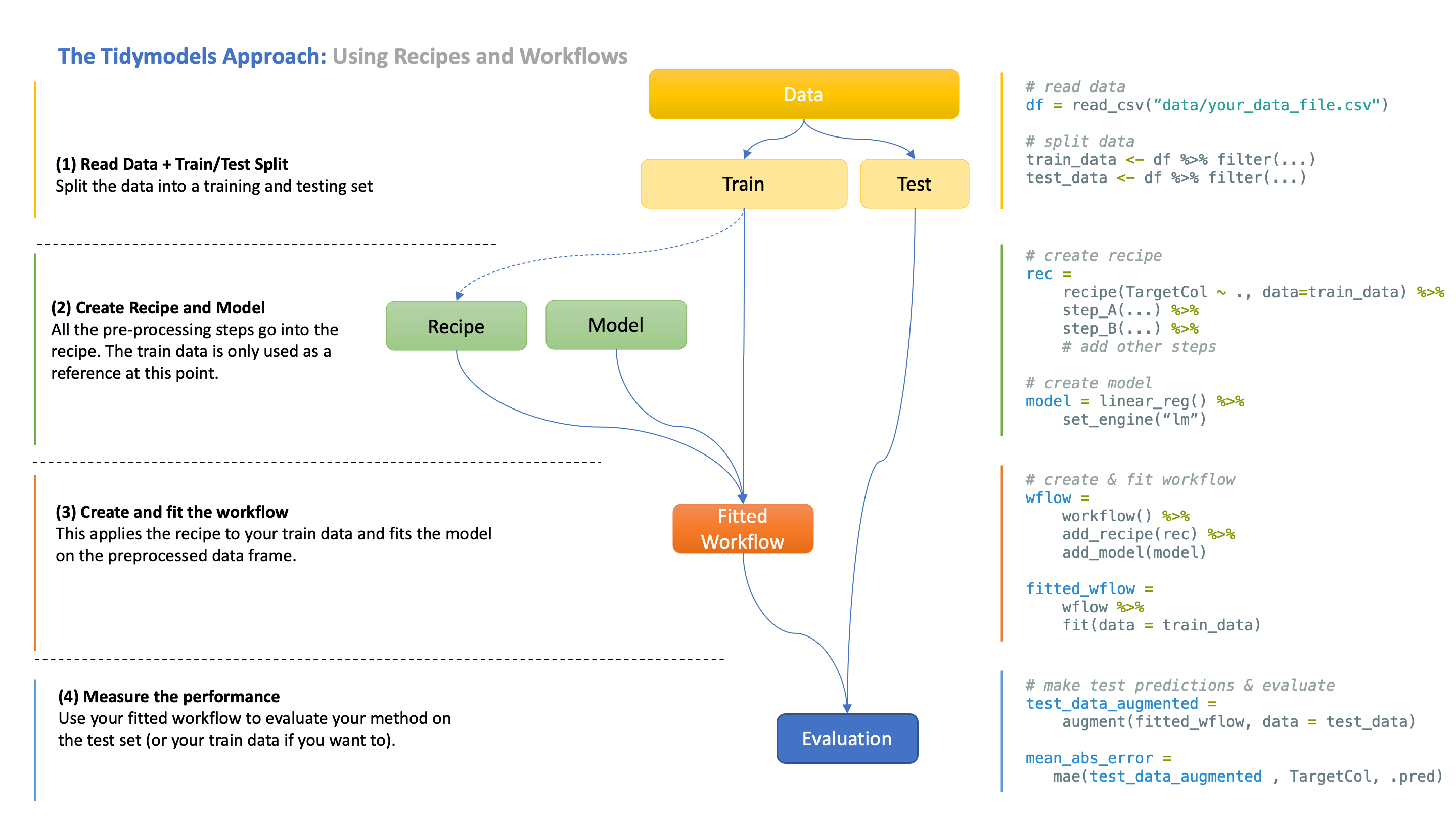
tidymodels approach(1) Split data into train and test sets
Why do we do this? Evaluating a model on the data it was trained in will give you a biased, overly optimistic estimate of the performance.
What you really want is for your model to generalise to new, unseen data. This is why you reserve a part of the data as a test set.
How is this split done? That depends! Sometimes, you randomly split the rows of the data set (e.g. 80% for training, 20% for testing), or sometimes you may want to split according to a different rule. In the current iteration of our course, DS202A - 2023/24 Autumn Term, for example, we have already split by region and time.
(2) Create recipe and workflow
What is a recipe? A recipe is a collection of instructions for how to process your data before it can be used for modelling.
Rarely is your data initially in an appropriate format for a model. Typically, you first need to clean the data, create new features, etc, and recipes make it easy to define those steps in a logical, reusable order. This is important as you always need to apply the same data pre-processing steps to both the training and the test data.
You may notice that the recipe already requires a reference to the train data. This is simply for the recipe to know the column names of the data, but no computations have occurred yet. The recipe is only a set of instructions, and things only really start to happen when you fit the workflow.
What is a workflow? The typical data science pipeline consists of processing and modelling, as the latter relies on the former.
A workflow combines those two steps so that they automatically run in this order when you use fit() or predict(). This saves you from writing additional code for running those steps individually.
(3) Fit the workflow
When you fit your workflow on the training data, your data is first processed according to the recipe, and then the processed data frame is fed into the model for training.
This workflow now also contains the fitted recipe and model, which you can extract if you need to.
(4) Evaluate the workflow
Finally, you can evaluate your workflow using the test data (or your train data if you wish).
What happens now is again a two-step procedure - your test data is first processed according to the recipe, and then your fitted model makes predictions.
Why did I say evaluate your “workflow” and not your “model”? The reason is that you actually always evaluate the whole pipeline. Your pre-processing choices affect the performance just like your model does, so it doesn’t really make sense to evaluate just one of them independently. They have to work jointly.
Conclusion
Using recipes and workflows often makes your code cleaner and easier to read. And generally, things work seamlessly together in the tidyverse.
Not always, however, which is why sometimes you can (and we do, too!) run into unexpected issues. One issue we have encountered after lab W04 of DS202A Autumn Term is that it’s advised not to create new predictors based on the outcome variable within recipes (we created a lag variable based on our outcome variable, yearly_change, if you remember). This causes augment(fitted_wflow, new_data = test_data) to fail.
A workaround is to extract the fitted recipe and model and apply them manually, which is very similar to what we also did in class.
# extract fitted recipe and model
fitted_rec = extract_recipe(fitted_wflow)
fitted_model = extract_fit_parsnip(fitted_wflow)
# manually bake test data using our recipe
test_data_baked = bake(fitted_rec, test_data)
# use fitted model and baked test data to make predictions
test_data_augmented = augment(fitted_model, new_data = test_data_baked)💡 Remember: if you avoid creating new variables or transformations based on the outcome, you shouldn’t have this problem, and you should be able to use the workflow directly to make predictions.
