✅ Week 02 Lab - Solutions
2024/25 Winter Term
This solution file follows the format of the Quarto file .qmd you had to fill in during the lab session.
Downloading the student solutions
Click on the below button to download the student notebook.
📋 Lab Tasks
🛠 Part 1: Data manipulation with pandas (45 min)
✨ pandas data frame attributes
A pandas data frame has a lot of attributes, several of which we shall explore:
shape: shows the number of rows and columns.index: prints the name of each row in the data frame.columns: prints the name of each column in the data frame.
🔧 pandas data frame methods
pandas data frames have a lot of methods! We will focus our attention on the following:
head/tail: shows the first / last n observations of a data frame.to_frame: converts a series (pandaswill automatically convert a data frame into a series if one variable is selected) to a data frame.unique: shows all unique values for qualitative features selected.value_counts: counts the number of times a unique value appears in a qualitative feature.query: keeps rows that conform to one or more logical criteria.reset_index: a subset of a data frame will keep the index of an old data frame. We use this method to change the index to one integer increments.filter: keeps columns that are included in a user-supplied list (via theitemsparameter).drop: drops columns that are included in a user-supplied list.rename: renames already existing columns based on a user-supplied dictionary.assign: creates a new variable based on alambdafunction.apply: applies alambdafunction across a set of variables.get_dummies: transforms qualitative features into a series of dummy features.groupby: perform grouped calculations within qualitative features.
⚙️ Setup
Import required libraries:
import numpy as np
import pandas as pd
from lets_plot import *
LetsPlot.setup_html()Import gapminder
gapminder = pd.read_csv("data/gapminder.csv")1.1: Printing pandas data frames
Let’s print gapminder.
# Printing the object itself
print(gapminder)
# Printing the first 5 rows
print(gapminder.head(5))
# Printing the last 5 rows
print(gapminder.tail(5))
country continent year lifeExp pop gdpPercap
0 Afghanistan Asia 1952 28.801 8425333 779.445314
1 Afghanistan Asia 1957 30.332 9240934 820.853030
2 Afghanistan Asia 1962 31.997 10267083 853.100710
3 Afghanistan Asia 1967 34.020 11537966 836.197138
4 Afghanistan Asia 1972 36.088 13079460 739.981106
... ... ... ... ... ... ...
1699 Zimbabwe Africa 1987 62.351 9216418 706.157306
1700 Zimbabwe Africa 1992 60.377 10704340 693.420786
1701 Zimbabwe Africa 1997 46.809 11404948 792.449960
1702 Zimbabwe Africa 2002 39.989 11926563 672.038623
1703 Zimbabwe Africa 2007 43.487 12311143 469.709298
[1704 rows x 6 columns]
country continent year lifeExp pop gdpPercap
0 Afghanistan Asia 1952 28.801 8425333 779.445314
1 Afghanistan Asia 1957 30.332 9240934 820.853030
2 Afghanistan Asia 1962 31.997 10267083 853.100710
3 Afghanistan Asia 1967 34.020 11537966 836.197138
4 Afghanistan Asia 1972 36.088 13079460 739.981106
country continent year lifeExp pop gdpPercap
1699 Zimbabwe Africa 1987 62.351 9216418 706.157306
1700 Zimbabwe Africa 1992 60.377 10704340 693.420786
1701 Zimbabwe Africa 1997 46.809 11404948 792.449960
1702 Zimbabwe Africa 2002 39.989 11926563 672.038623
1703 Zimbabwe Africa 2007 43.487 12311143 469.709298We can see the dimensions of our data by calling the shape attribute of gapminder.
gapminder.shape
(1704, 6)Here, we find that gapminder has 1,704 rows and 6 columns.
1.2: Data frames and series
We can see that gapminder has a list of countries and continents. We will explore continents further. To select only the continent column, we quote the variable name inside brackets next to gapminder.
gapminder["continent"]
0 Asia
1 Asia
2 Asia
3 Asia
4 Asia
...
1699 Africa
1700 Africa
1701 Africa
1702 Africa
1703 Africa
Name: continent, Length: 1704, dtype: objectYou can also reference variables using .:
gapminder.continent
0 Asia
1 Asia
2 Asia
3 Asia
4 Asia
...
1699 Africa
1700 Africa
1701 Africa
1702 Africa
1703 Africa
Name: continent, Length: 1704, dtype: object👉 NOTE: If you have variables separated by any white space or variables that contain special characters such as - or @, you can only use the brackets notation to select a column. The . (dot) notation can only be used for column names that are valid Python identifiers (e.g., no spaces, must start with a letter or underscore, and contain only alphanumeric characters or underscores).
This changes the data frame to a series. If you want the output to remain a data frame, however, you can use the to_frame method.
gapminder["continent"].to_frame()
continent
0 Asia
1 Asia
2 Asia
3 Asia
4 Asia
... ...
1699 Africa
1700 Africa
1701 Africa
1702 Africa
1703 Africa
[1704 rows x 1 columns]1.3: Finding / counting unique values
To find the names of all the continents we can use the unique method.
gapminder["continent"].unique()
array(['Asia', 'Europe', 'Africa', 'Americas', 'Oceania'], dtype=object)To see how many times each continent appears, we can use the value_counts method. We use reset index in order to turn continent into its own column.
gapminder["continent"].value_counts().reset_index()
continent count
0 Africa 624
1 Asia 396
2 Europe 360
3 Americas 300
4 Oceania 24❓Question: Do you see anything odd?
Yes, there are only 24 observations for Oceania, despite the fact that gapminder contains multiple measurements of the same country from 1952 to 2007.
1.4: Performing grouped calculations
Suppose we want to calculate average GDP per capita across time. We can use a combination of the groupby and mean methods from Pandas.
gapminder.groupby("year", as_index=False)["gdpPercap"].mean()
year gdpPercap
0 1952 3725.276046
1 1957 4299.408345
2 1962 4725.812342
3 1967 5483.653047
4 1972 6770.082815
5 1977 7313.166421
6 1982 7518.901673
7 1987 7900.920218
8 1992 8158.608521
9 1997 9090.175363
10 2002 9917.848365
11 2007 11680.0718201.5: Subsetting rows
Suppose we want to investigate as to why Oceania has only 24 observations (see Part 1.3), we can start by using the query method, which filters rows by one or more conditions.
gapminder_oceania = gapminder.query("continent == 'Oceania'")📝Task: Find the unique values of country in gapminder_oceania.
gapminder_oceania["country"].unique()
array(['Australia', 'New Zealand'], dtype=object)❓Question: Do you see the reason now?
Yes, this is because only Australia and New Zealand are included in Oceania. No other island nations are included.
1.6: Subsetting columns
Suppose we only want our data frame to include country, year and population. We can use the filter method in a Pandas data frame setting the items parameter equal to a list of feature names that we want to include.
gapminder.filter(items=["country","year","pop"])
country year pop
0 Afghanistan 1952 8425333
1 Afghanistan 1957 9240934
2 Afghanistan 1962 10267083
3 Afghanistan 1967 11537966
4 Afghanistan 1972 13079460
... ... ... ...
1699 Zimbabwe 1987 9216418
1700 Zimbabwe 1992 10704340
1701 Zimbabwe 1997 11404948
1702 Zimbabwe 2002 11926563
1703 Zimbabwe 2007 12311143
[1704 rows x 3 columns]Another option is to use the drop method which takes a list of features to drop. Here, we specify axis=1 which signifies columns, not rows (to specify rows, we set axis=0).
gapminder.drop(["lifeExp","continent","gdpPercap"],axis=1)
country year pop
0 Afghanistan 1952 8425333
1 Afghanistan 1957 9240934
2 Afghanistan 1962 10267083
3 Afghanistan 1967 11537966
4 Afghanistan 1972 13079460
... ... ... ...
1699 Zimbabwe 1987 9216418
1700 Zimbabwe 1992 10704340
1701 Zimbabwe 1997 11404948
1702 Zimbabwe 2002 11926563
1703 Zimbabwe 2007 12311143
[1704 rows x 3 columns]Yet another way to achieve the same result as the filter and drop methods we’ve just shown is the double square bracket subsetting:
gapminder[["country","year","pop"]]
country year pop
0 Afghanistan 1952 8425333
1 Afghanistan 1957 9240934
2 Afghanistan 1962 10267083
3 Afghanistan 1967 11537966
4 Afghanistan 1972 13079460
... ... ... ...
1699 Zimbabwe 1987 9216418
1700 Zimbabwe 1992 10704340
1701 Zimbabwe 1997 11404948
1702 Zimbabwe 2002 11926563
1703 Zimbabwe 2007 12311143
1704 rows × 3 columns👉 NOTE: If you have used pandas, you may have used the loc and iloc methods on data frames. These functions enable users to select both columns and rows in one function. While, in theory, this sounds great, these methods are computationally inefficient, so we advise that you do not use these methods and, instead, opt for query and filter or double square bracket subsetting.
1.7: Renaming columns
It is good practice to convert variable names to “snake case” whereby all characters are lower case and each word in the variable is separated by an underscore. To find the variable names expressed as an index, we call the columns attribute.
gapminder.columns
Index(['country', 'continent', 'year', 'lifeExp', 'pop', 'gdpPercap'], dtype='object')From there, we can amend the variable names using a dictionary where the key is the current variable name and the value is the variable name you would like it to be. We then use the rename method, setting the columns parameter equal to the dictionary created.
# Create a dictionary of variable names using snake case
snake_case_var_names = {
"country":"country",
"continent":"continent",
"year":"year",
"lifeExp": "life_exp",
"pop":"pop",
"gdpPercap":"gdp_per_cap"
}
# Set the columns attribute to this list
gapminder = gapminder.rename(columns=snake_case_var_names)
# Check the columns attribute
gapminder.columns
Index(['country', 'continent', 'year', 'life_exp', 'pop', 'gdp_per_cap'], dtype='object')1.8: Creating new variables
We know that Gross Domestic Product can be obtained from multiplying GDP per capita and population. To do this in Pandas, we simply insert * between the gdp_per_cap and pop columns found in gapminder.
gapminder["gdp"] = gapminder["gdp_per_cap"] * gapminder["pop"]
gapminder
country continent year life_exp pop gdp_per_cap \
0 Afghanistan Asia 1952 28.801 8425333 779.445314
1 Afghanistan Asia 1957 30.332 9240934 820.853030
2 Afghanistan Asia 1962 31.997 10267083 853.100710
3 Afghanistan Asia 1967 34.020 11537966 836.197138
4 Afghanistan Asia 1972 36.088 13079460 739.981106
... ... ... ... ... ... ...
1699 Zimbabwe Africa 1987 62.351 9216418 706.157306
1700 Zimbabwe Africa 1992 60.377 10704340 693.420786
1701 Zimbabwe Africa 1997 46.809 11404948 792.449960
1702 Zimbabwe Africa 2002 39.989 11926563 672.038623
1703 Zimbabwe Africa 2007 43.487 12311143 469.709298
gdp
0 6.567086e+09
1 7.585449e+09
2 8.758856e+09
3 9.648014e+09
4 9.678553e+09
... ...
1699 6.508241e+09
1700 7.422612e+09
1701 9.037851e+09
1702 8.015111e+09
1703 5.782658e+09
[1704 rows x 7 columns]After having calculated GDP, you may be interested in coding whether or not a country has above median GDP. We can turn where in Numpy into a function and create a new column using the assign method.
# Returns a boolean array if a quantitative feature is above median values
def is_above_median(var):
return np.where(var > np.median(var), True, False)
# Apply the function to GDP
gapminder[["country","year","gdp"]].assign(above_median = lambda x: is_above_median(x["gdp"]))
country year gdp above_median
0 Afghanistan 1952 6.567086e+09 False
1 Afghanistan 1957 7.585449e+09 False
2 Afghanistan 1962 8.758856e+09 False
3 Afghanistan 1967 9.648014e+09 False
4 Afghanistan 1972 9.678553e+09 False
... ... ... ... ...
1699 Zimbabwe 1987 6.508241e+09 False
1700 Zimbabwe 1992 7.422612e+09 False
1701 Zimbabwe 1997 9.037851e+09 False
1702 Zimbabwe 2002 8.015111e+09 False
1703 Zimbabwe 2007 5.782658e+09 False
[1704 rows x 4 columns]👉 NOTE: When using assign you can see that we use lambda x: followed by the function. All we are doing is using x as a placeholder for our data frame (country, year and gdp). In doing so, we can select the column we are interested in using to create our new boolean variable.
💁Tip: You may have noticed that we can string multiple methods together in Pandas. This is extremely useful, but you might find that your code will get too “long”. If you find this to be the case, you can use \ to spread your code over multiple lines. Here’s an example of how to do this with the above code:
gapminder[["country","year","gdp"]].\
assign(above_median = lambda x: is_above_median(x["gdp"]))
country year gdp above_median
0 Afghanistan 1952 6.567086e+09 False
1 Afghanistan 1957 7.585449e+09 False
2 Afghanistan 1962 8.758856e+09 False
3 Afghanistan 1967 9.648014e+09 False
4 Afghanistan 1972 9.678553e+09 False
... ... ... ... ...
1699 Zimbabwe 1987 6.508241e+09 False
1700 Zimbabwe 1992 7.422612e+09 False
1701 Zimbabwe 1997 9.037851e+09 False
1702 Zimbabwe 2002 8.015111e+09 False
1703 Zimbabwe 2007 5.782658e+09 False
[1704 rows x 4 columns]📝Task: This output is not very helpful. Try using some of the commands we have gone over to create a more useful data frame.
gapminder[["country","year","gdp"]].\
assign(above_median = lambda x: is_above_median(x["gdp"])).\
query("above_median == True").\
drop("above_median", axis=1).\
reset_index(drop=True)
country year gdp
0 Afghanistan 2007 3.107929e+10
1 Algeria 1952 2.272563e+10
2 Algeria 1957 3.095611e+10
3 Algeria 1962 2.806140e+10
4 Algeria 1967 4.143324e+10
.. ... ... ...
847 Vietnam 2007 2.081746e+11
848 Yemen, Rep. 1992 2.512511e+10
849 Yemen, Rep. 1997 3.351236e+10
850 Yemen, Rep. 2002 4.179396e+10
851 Yemen, Rep. 2007 5.065987e+10
[852 rows x 3 columns]1.9: Preparing data for scikit-learn, an example
📝Task: Filter the data to only include observations from 2007.
gapminder_07 = gapminder.query("year == 2007").reset_index(drop=True)
gapminder_07
country continent year life_exp pop gdp_per_cap \
0 Afghanistan Asia 2007 43.828 31889923 974.580338
1 Albania Europe 2007 76.423 3600523 5937.029526
2 Algeria Africa 2007 72.301 33333216 6223.367465
3 Angola Africa 2007 42.731 12420476 4797.231267
4 Argentina Americas 2007 75.320 40301927 12779.379640
.. ... ... ... ... ... ...
137 Vietnam Asia 2007 74.249 85262356 2441.576404
138 West Bank and Gaza Asia 2007 73.422 4018332 3025.349798
139 Yemen, Rep. Asia 2007 62.698 22211743 2280.769906
140 Zambia Africa 2007 42.384 11746035 1271.211593
141 Zimbabwe Africa 2007 43.487 12311143 469.709298
gdp
0 3.107929e+10
1 2.137641e+10
2 2.074449e+11
3 5.958390e+10
4 5.150336e+11
.. ...
137 2.081746e+11
138 1.215686e+10
139 5.065987e+10
140 1.493170e+10
141 5.782658e+09
[142 rows x 7 columns]📝Task: Create a list of numeric variables (life expectancy, population, GDP per capita, and GDP) and string variables (continent).
num_vars = ["life_exp","pop","gdp_per_cap","gdp"]
str_vars = ["continent"]📝Task: Subset the new data frame to only include these variables. Remember you can add elements to a list by using +.
gapminder_07 = gapminder_07.filter(items=num_vars+str_vars)
gapminder_07
<life_exp pop gdp_per_cap gdp continent
0 43.828 31889923 974.580338 3.107929e+10 Asia
1 76.423 3600523 5937.029526 2.137641e+10 Europe
2 72.301 33333216 6223.367465 2.074449e+11 Africa
3 42.731 12420476 4797.231267 5.958390e+10 Africa
4 75.320 40301927 12779.379640 5.150336e+11 Americas
.. ... ... ... ... ...
137 74.249 85262356 2441.576404 2.081746e+11 Asia
138 73.422 4018332 3025.349798 1.215686e+10 Asia
139 62.698 22211743 2280.769906 5.065987e+10 Asia
140 42.384 11746035 1271.211593 1.493170e+10 Africa
141 43.487 12311143 469.709298 5.782658e+09 Africa
[142 rows x 5 columns]📝Task: Apply normalise to numeric variables
# Create a function that normalises variables
def normalise(var):
return (var-var.mean())/var.std()
# Apply normalise function over all numeric variables
gapminder_07_normal = gapminder_07.filter(items=num_vars).\
apply(lambda x: normalise(x), axis=0)👉 NOTE: Normalising continuous features is good practice in machine learning, and becomes essential when dealing with algorithms that employ some kind of distance measure, such as principle components analysis.
Along with normalising features, we need to transform categorical features into one-hot encoded dummy (that is, 0 or 1) features. One-hot simply means that a reference category in a feature will not appear in the transformed data frame. To apply one-hot encoding, we can use get_dummies in Pandas.
gapminder_07_dummies = pd.get_dummies(gapminder_07["continent"],
columns=["continent"],
drop_first=True,
dtype=int)
gapminder_07_dummies
Americas Asia Europe Oceania
0 0 1 0 0
1 0 0 1 0
2 0 0 0 0
3 0 0 0 0
4 1 0 0 0
.. ... ... ... ...
137 0 1 0 0
138 0 1 0 0
139 0 1 0 0
140 0 0 0 0
141 0 0 0 0
[142 rows x 4 columns]After having transformed our continuous and categorical features, we can concatenate the two into a new data frame.
gapminder_07_cleaned = pd.concat([gapminder_07_normal, gapminder_07_dummies], axis=1)
gapminder_07_cleaned
life_exp pop gdp_per_cap gdp Americas Asia Europe \
0 -1.919936 -0.082178 -0.832468 -0.288250 0 1 0
1 0.779886 -0.273813 -0.446584 -0.295646 0 0 1
2 0.438463 -0.072401 -0.424318 -0.153810 0 0 0
3 -2.010799 -0.214066 -0.535216 -0.266522 0 0 0
4 0.688525 -0.025195 0.085483 0.080659 1 0 0
.. ... ... ... ... ... ... ...
137 0.599815 0.279371 -0.718394 -0.153254 0 1 0
138 0.531315 -0.270983 -0.672999 -0.302674 0 1 0
139 -0.356947 -0.147739 -0.730898 -0.273324 0 1 0
140 -2.039541 -0.218635 -0.809402 -0.300559 0 0 0
141 -1.948180 -0.214807 -0.871728 -0.307533 0 0 0
Oceania
0 0
1 0
2 0
3 0
4 0
.. ...
137 0
138 0
139 0
140 0
141 0
[142 rows x 8 columns]👉 NOTE: These kinds of transformations and concatenations will be employed a lot during this course, so please be sure to get used to them.
As a final (optional) step, we can convert our Pandas data frame to a Numpy array by employing the to_numpy method.
gapminder_07_cleaned.to_numpy()
array([[-1.91993566, -0.08217844, -0.8324684 , ..., 1. ,
0. , 0. ],
[ 0.77988582, -0.27381326, -0.446584 , ..., 0. ,
1. , 0. ],
[ 0.43846339, -0.07240145, -0.42431811, ..., 0. ,
0. , 0. ],
...,
[-0.35694651, -0.14773926, -0.73089796, ..., 1. ,
0. , 0. ],
[-2.03954118, -0.21863487, -0.8094021 , ..., 0. ,
0. , 0. ],
[-1.94818045, -0.21480678, -0.87172762, ..., 0. ,
0. , 0. ]])🏆Challenge: Which countries had above average life expectancy in 1952?
Try these steps: 1. Define a function that returns a boolean if a value in a feature exceeds the average value. 2. Include only the year 1952 by using query. 3. Subset the data frame to only include country and life expectancy using filter. 4. Create a new boolean variable using the user defined function using assign. 5. Include all rows with above average life expectancy using query. 6. Subset the data frame to only include country using square bracket indexing. 7. Pull the unique values into an array using unique.
# Returns a boolean array based on whether a quantitative feature has above average values
def is_above_average(var):
return np.where(var > np.mean(var), True, False)
# Answer to be provided
gapminder.\
query("year == 1952").\
filter(items=["country","life_exp"]).\
assign(above_average = lambda x: is_above_average(x["life_exp"])).\
query("above_average == True")\
["country"].\
unique()
array(['Albania', 'Argentina', 'Australia', 'Austria', 'Bahrain',
'Belgium', 'Bosnia and Herzegovina', 'Brazil', 'Bulgaria',
'Canada', 'Chile', 'Colombia', 'Costa Rica', 'Croatia', 'Cuba',
'Czech Republic', 'Denmark', 'Finland', 'France', 'Germany',
'Greece', 'Hong Kong, China', 'Hungary', 'Iceland', 'Ireland',
'Israel', 'Italy', 'Jamaica', 'Japan', 'Korea, Dem. Rep.',
'Kuwait', 'Lebanon', 'Mauritius', 'Mexico', 'Montenegro',
'Netherlands', 'New Zealand', 'Norway', 'Panama', 'Paraguay',
'Poland', 'Portugal', 'Puerto Rico', 'Reunion', 'Romania',
'Serbia', 'Singapore', 'Slovak Republic', 'Slovenia', 'Spain',
'Sri Lanka', 'Sweden', 'Switzerland', 'Taiwan', 'Thailand',
'Trinidad and Tobago', 'United Kingdom', 'United States',
'Uruguay', 'Venezuela'], dtype=object)📋 Part 2: Data visualisation with lets_plot(45 min)
They say a picture paints a thousand words, and we are, more often than not, inclined to agree with them (whoever “they” are)! Thankfully, we can use perhaps the most widely used package, lets_plot (click here for documentation) to paint such pictures or (more accurately) build such graphs.
2.1 Histograms
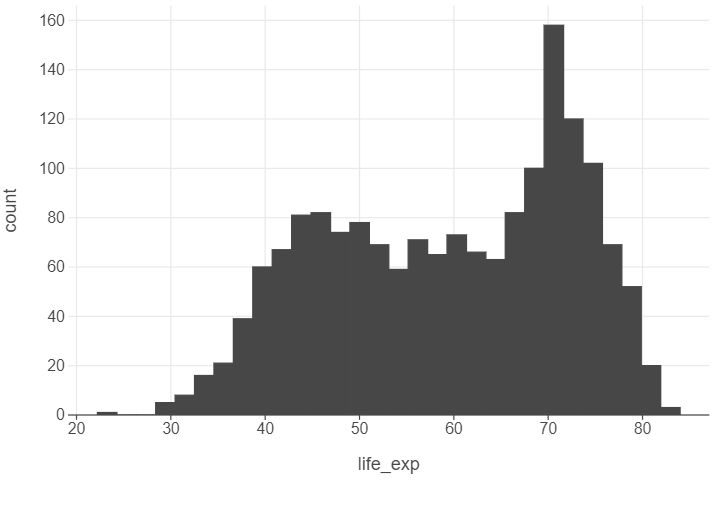
Suppose we want to plot the distribution of an outcome of interest. We can use a histogram to plot the distribution of life expectancy in gapminder.
📝 Note: Step-by-step
If you are not familiar with using lets_plot for plotting, the line by line construction of the plot below might be useful. The + operator is used to add layers to the plot.
(
ggplot(gapminder, aes("life_exp")) +
geom_histogram()
)
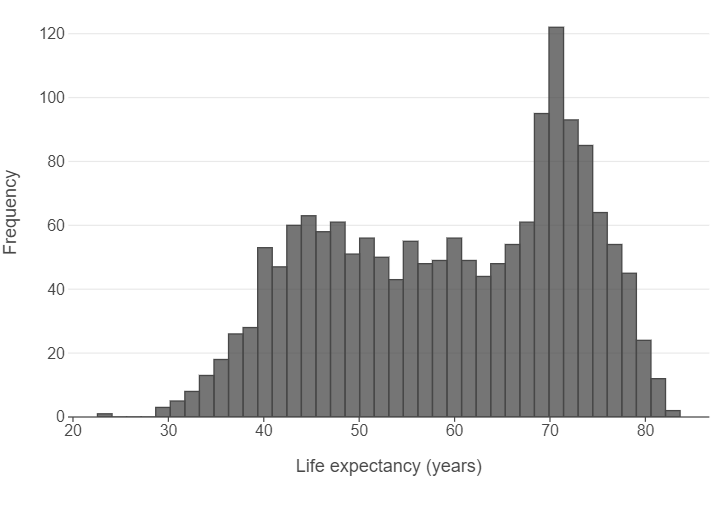
👥 WORK IN PAIRS/GROUPS:
Change a few parameters in the geom_histogram layer. - Set bins to 40. - Set alpha to 0.75.
Afterwards, - Specify panel_grid_major_x=element_blank() in the theme function. - Edit the x and y axes using labs.
(
ggplot(gapminder, aes("life_exp")) +
geom_histogram(alpha = 0.75, bins = 40) +
theme(panel_grid_major_x=element_blank()) +
labs(x = "Life expectancy (years)", y = "Frequency")
)
2.2 Bar graphs
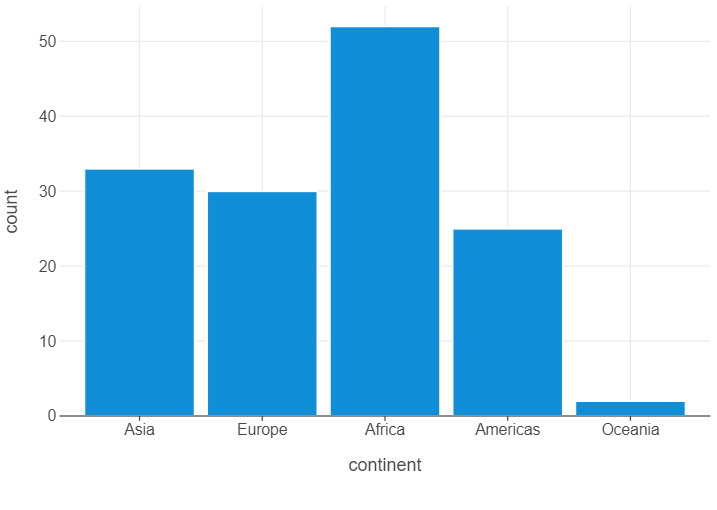
We counted the number of countries in each continent that are in gapminder. We will do something similar, using a subset of the data in 1997. We can use lets_plot to create a bar graph. We first need to specify what we want on the x and y axes, which constitute the first and second argument of the aes function.
gapminder_97 = gapminder.query("year == 1997")
(
ggplot(gapminder_97,aes("continent")) +
geom_bar()
)
👥 WORK IN PAIRS/GROUPS:
- Create a new data frame that counts the number of observations for each continent.
- Set
stat="identity"ingeom_bar. - Specify
panel_grid_major_x=element_blank()in thethemefunction. - Edit the x and y axes using
labs.
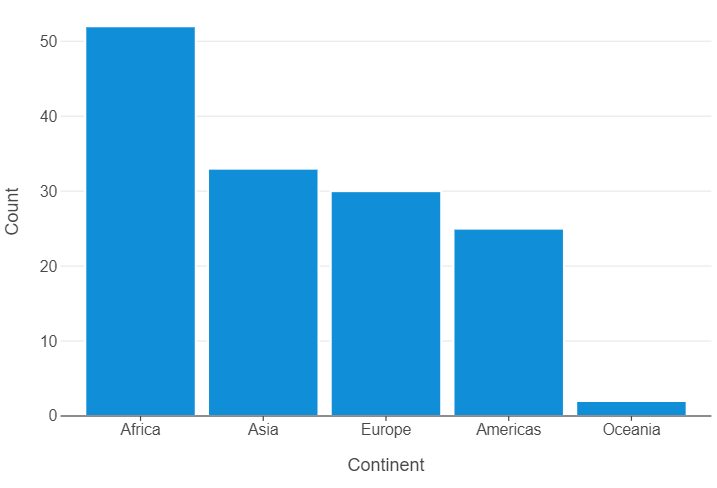
gapminder_97_count = gapminder_97.value_counts("continent").reset_index()
(
ggplot(gapminder_97_count, aes("continent","count")) +
geom_bar(stat="identity") +
theme(panel_grid_major_x=element_blank()) +
labs(x="Continent",y="Count")
)
👉 NOTE: Don’t like vertical bars? Well, we can also create horizontal bars too by specifying orientation="y" in geom_bar. Remember to switch “count” and “continent”.
2.3 Scatter plots
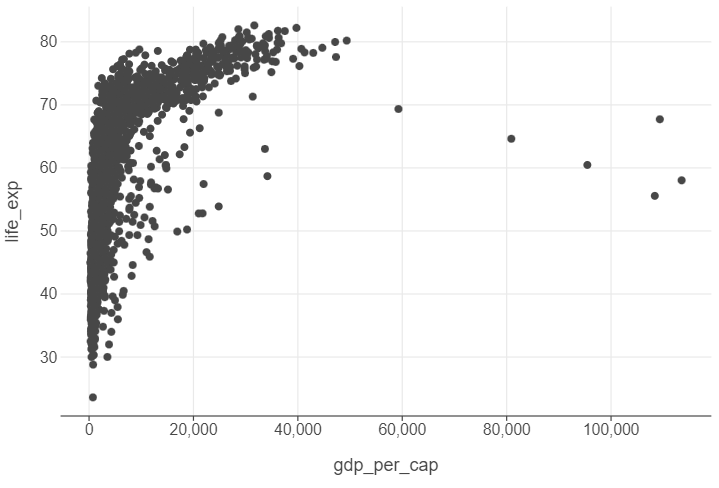
Scatter plots are the best way to graphically summarise the relationship between two quantitative features. Suppose we wanted to visualise the “effect” of GDP per capita on life expectancy. We can use a scatter plot to achieve this.
(
ggplot(gapminder, aes("gdp_per_cap","life_exp")) +
geom_point()
)
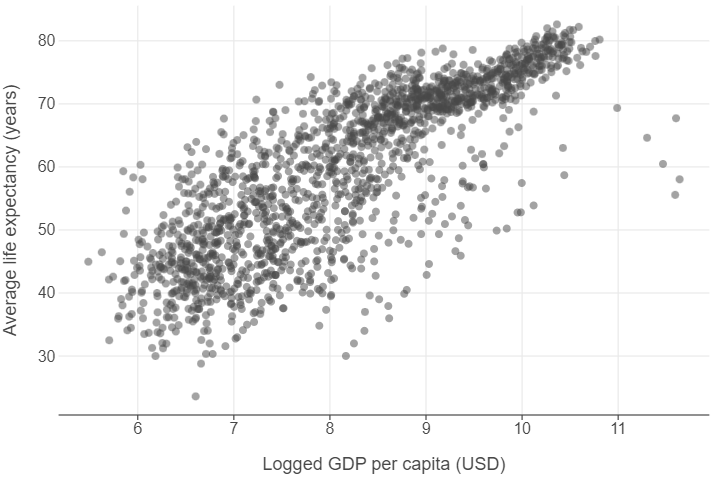
👥 WORK IN PAIRS/GROUPS:
- Create a new feature that takes the natural log of GDP per capita.
- Specify
alpha=0.5ingeom_point. - Edit the x and y axes using
labs.
gapminder["log_gdp_per_cap"] = np.log(gapminder["gdp_per_cap"])
(
ggplot(gapminder, aes("log_gdp_per_cap","life_exp")) +
geom_point(alpha=0.5) +
labs(x="Logged GDP per capita (USD)", y = "Average life expectancy (years)")
)
2.4 Line plots
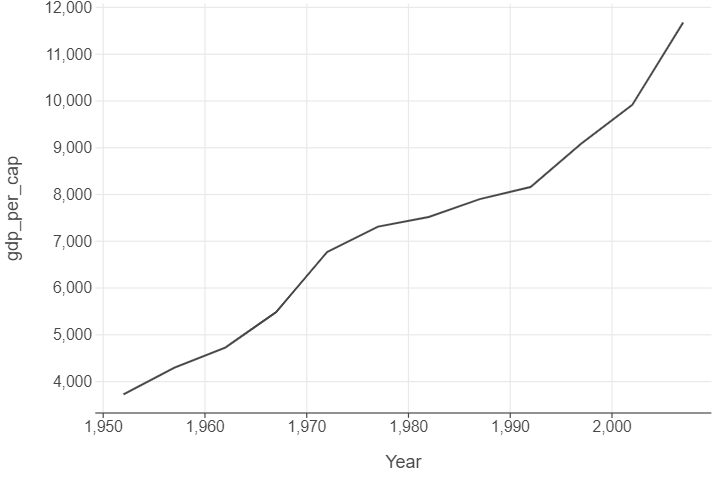
Line plots can be used to track parameters of interest over time. We will do this by calculating then plotting average GDP per capita for each year.
gdp_per_cap_by_year = gapminder.groupby("year", as_index=False)["gdp_per_cap"].mean()
(
ggplot(gdp_per_cap_by_year, aes("year", "gdp_per_cap")) +
geom_line() +
labs(x = "Year")
)
👥 WORK IN PAIRS/GROUPS:
- Change
linetype="dotted"ingeom_line. - Add a
geom_pointandgeom_arealayer. - Change the
breaksparameter inscale_x_continuousto an array from 1952 to 2007 by increments of 10. - Specify
panel_grid_major_x=element_blank()in thethemefunction. - Edit the x and y axes using
labs.
(
ggplot(gdp_per_cap_by_year, aes("year", "gdp_per_cap")) +
geom_line(linetype="dotted") +
geom_point() +
geom_area() +
scale_x_continuous(breaks=np.arange(1952,2007,10)) +
theme(panel_grid_major_x=element_blank()) +
labs(x = "Year", y = "Average GDP per capita (USD)")
)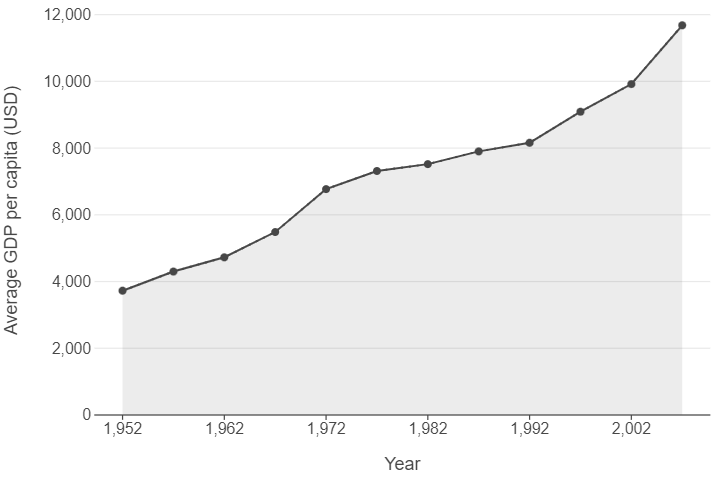
👉 NOTE: We think this line plot works in this context, and that it is useful to introduce you to this style of charting progress over time. However, for others contexts filling the area under the line may not work, so do bear this in mind when making decisions!
2.5 Box plots
Box plots are an excellent way to summarise the distribution of values across different strata over key quantities of interest (e.g. median, interquartile range). Suppose we want to show the distribution of populations by continent in gapminder in 1982 (excluding Oceania). We can use box plots to graphically illustrate this.
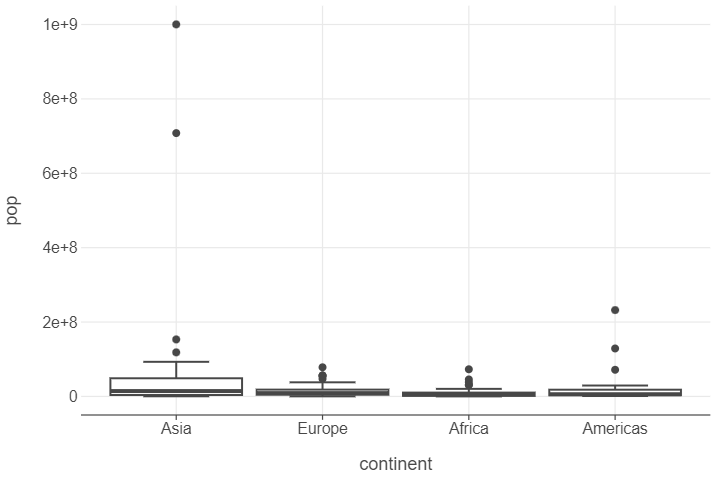
gapminder_oceania_82 = gapminder.query("continent != 'Oceania' & year == 1982")
(
ggplot(gapminder_oceania_82, aes("continent", "pop")) +
geom_boxplot()
)
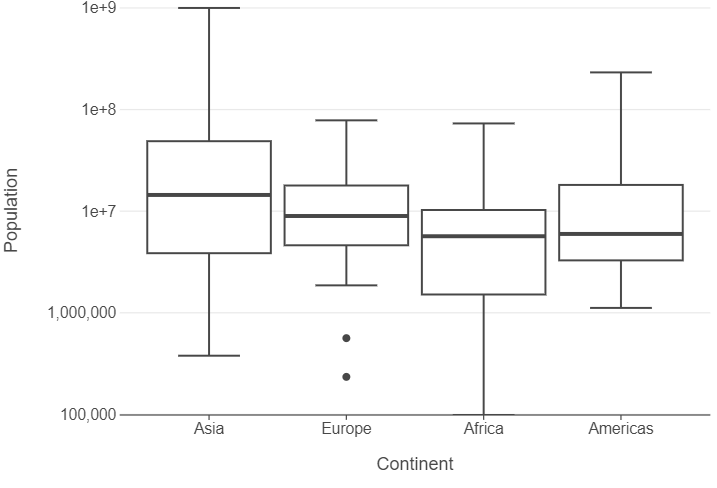
👥 WORK IN PAIRS/GROUPS:
- Add
scale_y_log10to the plot. - Specify
panel_grid_major_x=element_blank()in thethemefunction. - Edit the x and y axes using
labs.
(
ggplot(gapminder_oceania_82, aes("continent", "pop")) +
geom_boxplot() +
scale_y_log10() +
theme(panel_grid_major_x=element_blank()) +
labs(x="Continent", y="Population")
)
2.6 Density plots
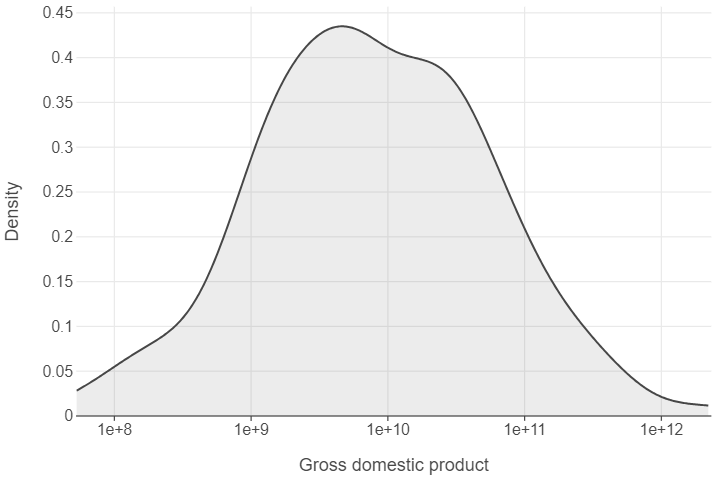
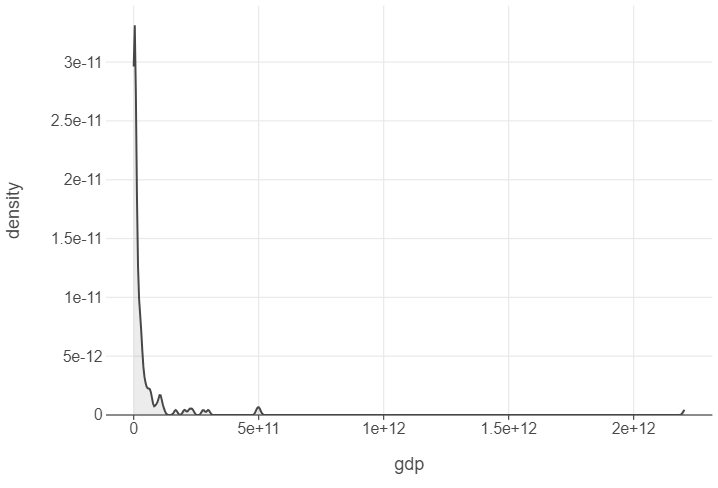
Finally, we can understand the distribution of continuous features using density plots. Here’s an example that looks at the distribution of GDP in 1952.
gapminder_52 = gapminder.query("year == 1952")
(
ggplot(gapminder_52, aes("gdp")) +
geom_density()
)
👥 WORK IN PAIRS/GROUPS:
- Add
scale_x_log10to the plot. - Specify
panel_grid_major_x=element_blank()in thethemefunction. - Edit the x and y axes using
labs.
gapminder_52 = gapminder.query("year == 1952")
(
ggplot(gapminder_52, aes("gdp")) +
geom_density() +
scale_x_log10() +
labs(x="Gross domestic product", y="Density")
)