✅ Week 04 Lab - Solutions
aka: The one where I get complain about the recipes package
You will find that I got really annoyed with the recipes package in this lab. I am rethinking my position on it. I might simply abandon it in this course and return to the old way of doing things, with pure dplyr and tidyr. I will let you know in the next lecture. While this is definitely annoying, it is also a picture of how coding works in real life! You need to pay close attention to the details, and sometimes, the brandished solution might not work, and we have to adapt. Well, that’s how we learn, I guess!
Import required libraries:
# Tidyverse packages we will use
library(dplyr)
library(tidyr)
library(readr)
library(lubridate)
# Tidymodel packages we will use
library(yardstick) # this is where metrics like mae() are
library(parsnip) # This is where models like linear_reg() are
library(recipes) # You will learn about this today
library(workflows) # You will learn about this todayRead the data set:
It is the same data set you used in the previous lab (as well as in the first formative assignment):
# Modify the filepath if needed
filepath <- "data/UK-HPI-full-file-2023-06.csv"
uk_hpi <- read_csv(filepath)Part III - Practice with recipes and workflows (40 min)
We aim to work out a recipe from scratch directly from the uk_hpi data set.
Task 1:
Question: Can you replicate the filtering steps we applied to
uk_hpifor England, but this time, use a recipe for the same tasks?
- Employ a
step_filter()to limit the records to those from 2005 and beyond.- Use
step_mutate()to format theDatecolumn correctly.- Set all variables other than
SalesVolumeasidvariables.- Generate the lagged variable of
SalesVolumeas done previously.- Eliminate rows with missing values as done previously.
If you followed our instructions in the lab, you would have arrived at the following:
rec <-
recipe(SalesVolume ~ ., data = uk_hpi) %>%
update_role(-SalesVolume, new_role = "id") %>%
step_mutate(Date = dmy(Date), skip=FALSE) %>%
step_filter(Date >= ymd("2005-01-01"), skip=FALSE) %>%
step_arrange(RegionName, Date) %>%
step_lag(SalesVolume, lag = 1, skip=FALSE) %>%
step_naomit(-c(SalesVolume, lag_1_SalesVolume)) %>%
prep() Note: I moved the update_role() to the top of the recipe, as I found it easier to use a negative selection before any other steps.
I was unaware of this until I started preparing these solutions: we should not add any step_ that involves the outcome variable (in our case, SalesVolume) in the recipes. Apparently, we should treat it only outside the recipe. I will do a bit more of digging into this and share with you later. Live and learn…
Kudos to @astoeffelbauer for pointing this out to me. In his words:
“Read here and here why, which makes sort of sense but is still a little annoying…”
I agree! I’m already annoyed that we have to add skip=FALSE to some steps, and now this… The tidymodels developers will hear from me soon!
So, what should we do then?
We should create the lagged variable outside the recipe, using the usual dplyr transformations, and then add it to the data set. It’s a little like how we did it before learning about recipes:
filepath <- "data/UK-HPI-full-file-2023-06.csv"
uk_hpi <-
read_csv(filepath) %>%
mutate(Date = dmy(Date)) %>%
group_by(RegionName) %>%
arrange(Date) %>%
mutate(lag_1_SalesVolume = lag(SalesVolume, 1)) %>% # I decided to use the same naming convention as in the recipe
ungroup()
# Now my recipe does not need to include anything about the outcome variable
rec <-
recipe(SalesVolume ~ ., data = uk_hpi) %>%
update_role(-c(SalesVolume, lag_1_SalesVolume), new_role = "id") %>%
step_filter(Date >= ymd("2005-01-01"), skip=FALSE) %>%
step_naomit(-c(SalesVolume, lag_1_SalesVolume)) %>%
prep() Task 2:
Using this recipe, train a model that uses as training data only the records of England, Scotland and Wales. Calculate the Mean Absolute Error (MAE) of the training data. Then, respond: how does it compare to the MAE of the model trained on all UK countries (Part II)?
Here, I will choose to split the data into training and testing sets before I apply the recipe:
uk_hpi_train <- uk_hpi %>% filter(RegionName %in% c("England", "Scotland", "Wales"))
uk_hpi_test <- uk_hpi %>% filter(RegionName == "Northern Ireland")Now, I can train a model on this data set:
# Because I am using recipes, I don't need to specify the formula
lm_specification <-
linear_reg() %>%
set_engine("lm")
# Create a workflow
lm_workflow <-
workflow() %>%
add_model(lm_specification) %>%
add_recipe(rec)
# Train the model using the workflow and the training data set
lm_fit <-
lm_workflow %>%
fit(uk_hpi_train)I can now use the model to make predictions on the training data set:
lm_fit %>%
extract_fit_parsnip() %>%
augment(rec %>% bake(uk_hpi_train)) %>%
group_by(RegionName) %>%
mae(truth = SalesVolume, estimate = .pred)This time, the MAE was larger for all countries involved:
# A tibble: 3 x 4
RegionName .metric .estimator .estimate
<fct> <chr> <chr> <dbl>
1 England mae standard 10164.
2 Scotland mae standard 1533.
3 Wales mae standard 1423.Task 3:
Now, let’s think of Northern Ireland records as a testing set. Predict SalesVolume for Northern Ireland using the model you fitted in Step 2. Calculate the MAE of the predictions. What do you make of it?
lm_fit %>%
extract_fit_parsnip() %>%
augment(rec %>% bake(uk_hpi_test)) %>%
group_by(RegionName) %>%
mae(truth = SalesVolume, estimate = .pred)Which is way worse than the MAE of the model trained in all UK countries:
# A tibble: 1 x 4
RegionName .metric .estimator .estimate
<fct> <chr> <chr> <dbl>
1 Northern Ireland mae standard 1494.This is more expected. After all, the previous model had complete information about the data set, while this one was trained on a subset of the data. I will comment more on this in the next task.
Task 4:
Write a function called
plot_residuals()that takes a fitted workflow model and a data frame in its original form (that is, before any pre-processing) and plot the residuals against the fitted values. The function should return a ggplot object.
plot_residuals <- function(wflow_model, data) {
# Extract the recipe from the workflow
rec <- wflow_model %>% extract_recipe()
# Apply the recipe to the data
plot_df <-
wflow_model %>%
extract_fit_parsnip() %>%
augment(rec %>% bake(data))
g <- ggplot(plot_df, aes(x=.pred, y=.resid, fill=RegionName, color=RegionName)) +
geom_point() +
geom_hline(yintercept = 0, linetype="dashed") +
labs(x="Fitted values", y="Residuals") +
facet_wrap(~RegionName, nrow=1, scales="free_x") +
theme_bw()
return(g)
}Then I call:
plot_residuals(lm_fit, uk_hpi %>% filter(RegionName %in% c("England", "Scotland", "Wales", "Northern Ireland")))Which makes me annoyed at recipes again! This is difficult to debug, but apparently, the recipe converts character columns to factors internally – even those that are not predictors. Therefore, when I add ‘Northern Ireland’ to the list of countries above, the bake() function ends up converting the rows that contain ‘Northern Ireland’ to NA, and we lose the RegionName information. This is why the final plot does not have Northern Ireland in the title, even though it is all Northern Ireland data!!! Bad tidymodels!
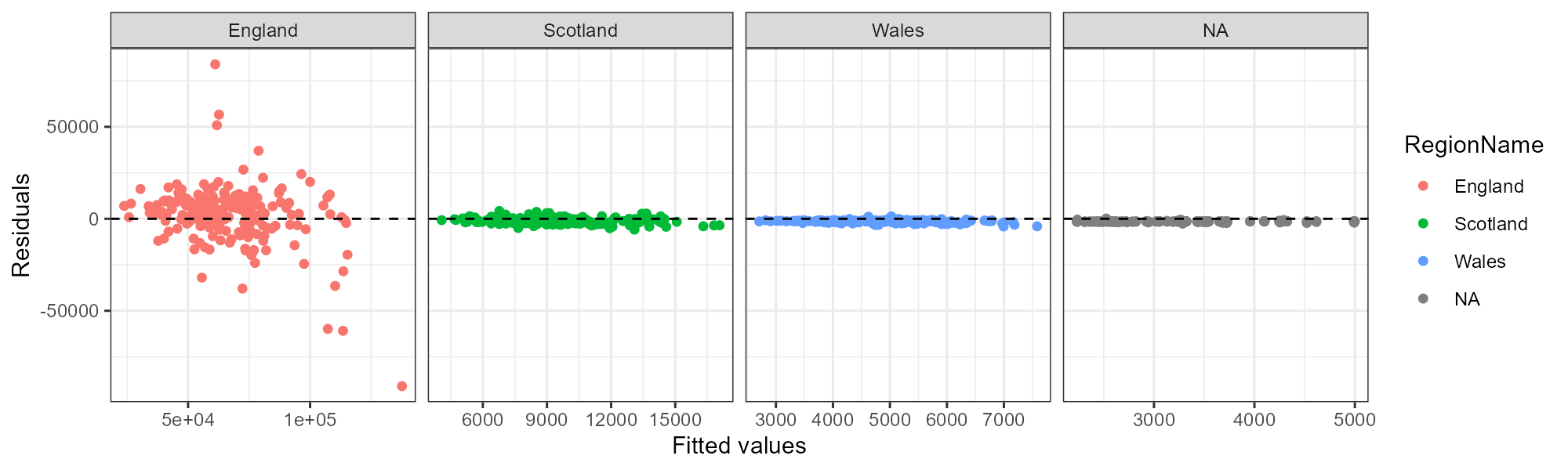
Anyways, what can we learn from this plot? Notice that I let the X-axis free to adjust to each of the individual plots. After all, the number of units sold in each country is very different. But, I kept the Y-axis fixed to compare the residuals across countries.
The main takeaway is that England is the most difficult subset of this data to model. The residuals are all over the place, and this model is probably not the absolute best we can do. What do you think we should do next?
