library('ISLR2') # for the data
library('tidyverse') # to use things like the pipe (%>%)
library('e1071') # for SVM model
library('tidymodels') # for model tuning, cross-validation etc.
# Vanity packages:
library('GGally') # for pretty correlation plot
library('ggsci') # for pretty plot colours
library('cvms') # for pretty confusion matrix plots💻 Week 08 - Lab Roadmap (90 min)
DS202 - Data Science for Social Scientists
Context
This lab session draws on 🗓️ Week 04 lecture/workshop and 🗓️ Week 05 lecture content.
It also reuses elements of tidymodels introduced in previous labs where we used functions from library(broom) or library(rsample). These packages are part of tidymodels. For the record, tidymodel’s ‘Get Started’ tutorials are really good.
Download this page as an RMarkdown file from Moodle.
Topics
More specifically, these are the things we will explore or revisit:
- Difference between Classification vs Regression (🗓️ Week 03 lecture)
- k-fold cross-validation
- Support Vector Machine (SVM)
- Hyperparameters
Setup
Setup
💡 Some of you have mentioned that your R version cannot handle tidymodels. We recommend you update R to version 4.2.0 or above.
Packages you will need
The Data (10 min)
The Data (10 min)
🍊Orange Juice
This week we will use a different ISLR2 dataset: OJ . We will perform a classification task with the goal to predict the Purchase column.
The data contains 1070 purchases where the customer either purchased Citrus Hill or Minute Maid Orange Juice. A number of characteristics of the customer and product are recorded.
ISLR2::OJ %>% head() Purchase WeekofPurchase StoreID PriceCH PriceMM DiscCH DiscMM SpecialCH
1 CH 237 1 1.75 1.99 0.00 0.0 0
2 CH 239 1 1.75 1.99 0.00 0.3 0
3 CH 245 1 1.86 2.09 0.17 0.0 0
4 MM 227 1 1.69 1.69 0.00 0.0 0
5 CH 228 7 1.69 1.69 0.00 0.0 0
6 CH 230 7 1.69 1.99 0.00 0.0 0
SpecialMM LoyalCH SalePriceMM SalePriceCH PriceDiff Store7 PctDiscMM
1 0 0.500000 1.99 1.75 0.24 No 0.000000
2 1 0.600000 1.69 1.75 -0.06 No 0.150754
3 0 0.680000 2.09 1.69 0.40 No 0.000000
4 0 0.400000 1.69 1.69 0.00 No 0.000000
5 0 0.956535 1.69 1.69 0.00 Yes 0.000000
6 1 0.965228 1.99 1.69 0.30 Yes 0.000000
PctDiscCH ListPriceDiff STORE
1 0.000000 0.24 1
2 0.000000 0.24 1
3 0.091398 0.23 1
4 0.000000 0.00 1
5 0.000000 0.00 0
6 0.000000 0.30 0To understand what each variable represent, open the R Console, type the following and hit ENTER:
?ISLR2::OJWhich variables can help us distinguish the two different brands?
To simplify our plots later on, let’s focus on just two predictors:
plot_df <- ISLR2::OJ %>% select(Purchase, LoyalCH, PriceDiff)
g = (
ggpairs(plot_df, aes(colour=Purchase))
# Customizing the plot
+ scale_colour_startrek()
+ scale_fill_startrek()
+ theme_bw()
)
g 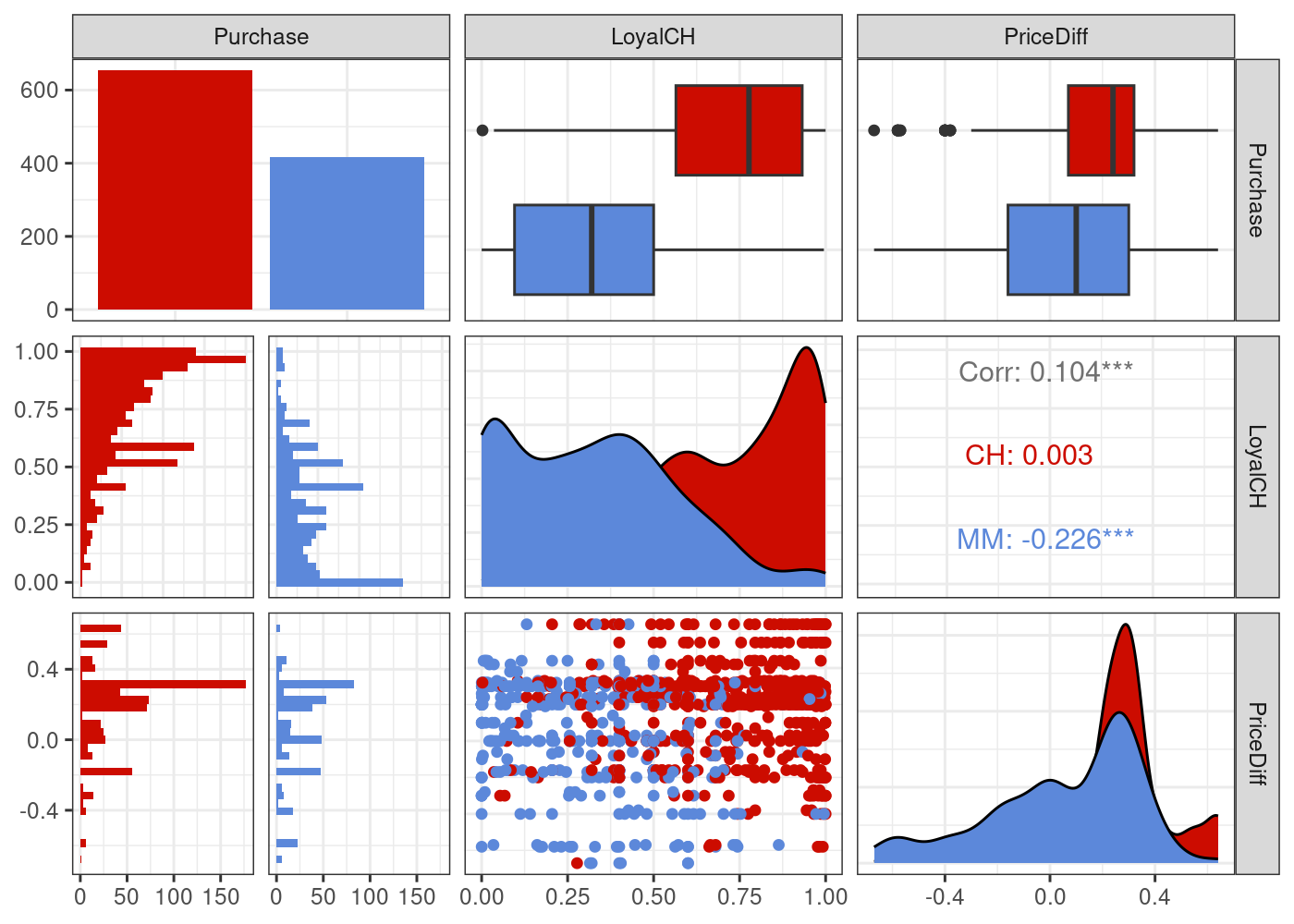
📈 Stock Market
We will also use the Smarket dataset from the ISLR2 package. We will perform a regression task with the goal to predict the percentage of return of the S&P 500 stock index on any given day, as represented by the Today column.
Daily percentage returns for the S&P 500 stock index between 2001 and 2005.
ISLR2::Smarket %>% head() Year Lag1 Lag2 Lag3 Lag4 Lag5 Volume Today Direction
1 2001 0.381 -0.192 -2.624 -1.055 5.010 1.1913 0.959 Up
2 2001 0.959 0.381 -0.192 -2.624 -1.055 1.2965 1.032 Up
3 2001 1.032 0.959 0.381 -0.192 -2.624 1.4112 -0.623 Down
4 2001 -0.623 1.032 0.959 0.381 -0.192 1.2760 0.614 Up
5 2001 0.614 -0.623 1.032 0.959 0.381 1.2057 0.213 Up
6 2001 0.213 0.614 -0.623 1.032 0.959 1.3491 1.392 Upplot_df <- ISLR2::Smarket %>% select(Today, Volume, Lag1, Lag2)
g = (
ggpairs(plot_df)
# Customizing the plot
+ scale_colour_startrek()
+ scale_fill_startrek()
+ theme_bw()
)
g 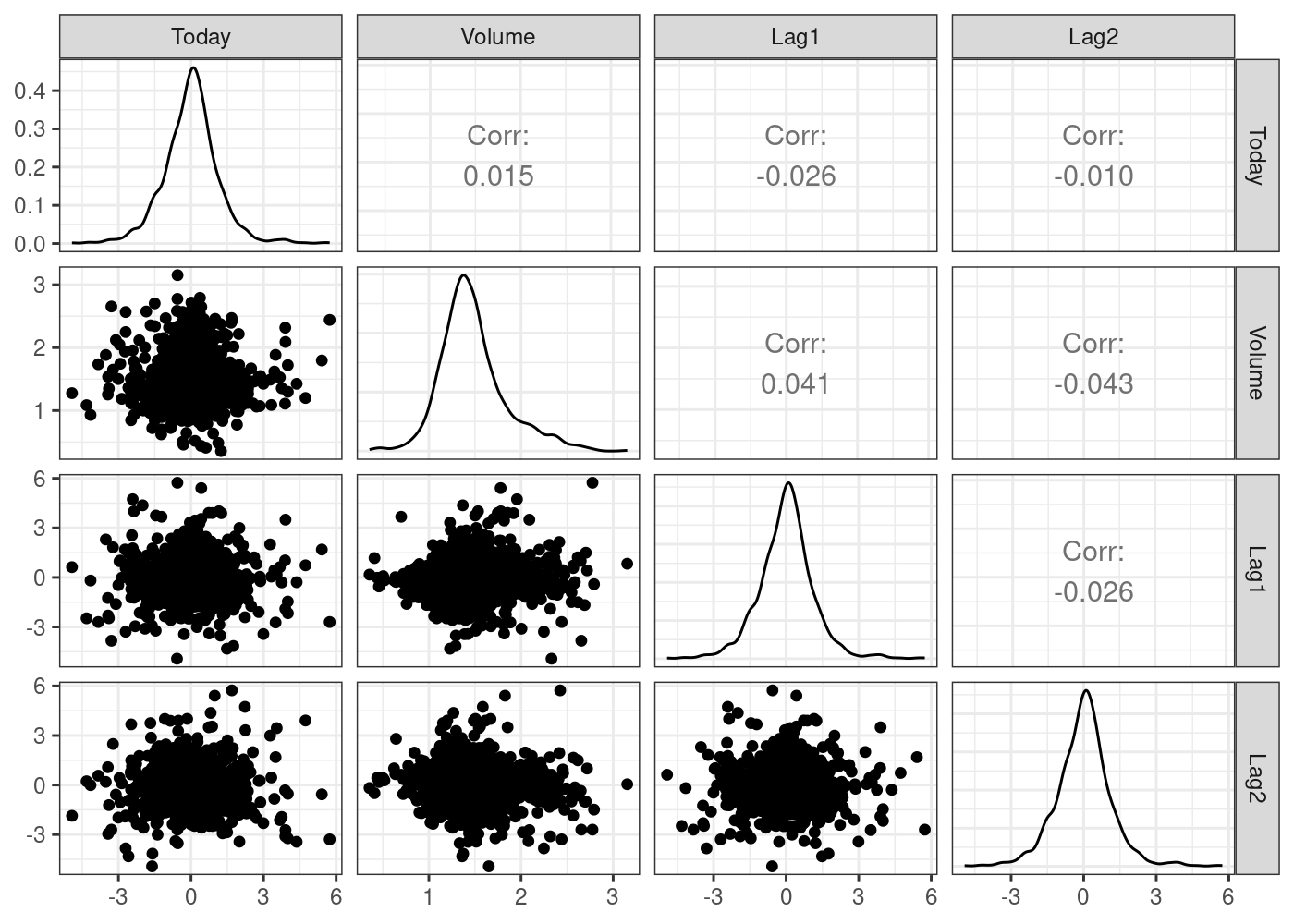
To understand what each variable represent, open the R Console, type the following and hit ENTER:
?ISLR2::SmarketStep 1: SVM models for classification (30 min)
Step 1: SVM models for classification (30 min)
SVM stands for Support Vector Machines. Revisit 🗓️ Week 05 lecture or Chapter 9 of our textbook to understand more about this algorithm.
R does not come with SVM, so we need to import it from a library. Let’s start with the function svm we used in the Week 05 lecture, imported from the e1071 package. Here are a few things to know about the svm function:
- The
svmcommand is largely similar to other commands such aslmandglmin that the first parameter is an R formula and the second is the data set. - There are a few other options, but we will focus on specifying a radial kernel using
kernel = 'radial'. - We can specify the type of machine learning task we are performing. Since we are doing classification, we use the option
type = 'C-classification'.
Step 1.1: Train a SVM model
filtered_data <- ISLR2::OJ %>% select(Purchase, LoyalCH, PriceDiff)
orange_svm_model <- svm(Purchase ~ .,
data=filtered_data,
kernel='radial',
type='C-classification')
orange_svm_model
Call:
svm(formula = Purchase ~ ., data = filtered_data, kernel = "radial",
type = "C-classification")
Parameters:
SVM-Type: C-classification
SVM-Kernel: radial
cost: 1
Number of Support Vectors: 427🎯 ACTION POINT: What does the ‘Number of Support Vectors’ represent?
your text here
🎯 ACTION POINT: What would happened if we changed kernel to kernel="linear"?
your text here
Step 1.2: Goodness-of-Fit of the SVM
Let’s investigate how well our model fits the data. Let’s reuse the model we trained (orange_svm_model) and predict the same samples we used to train it. To avoid modifying our original dataframe, let’s save the output of the prediction in an auxiliary df (plot_df):
plot_df <-
filtered_data %>%
mutate(class_pred = predict(orange_svm_model, newdata = .))
head(plot_df)Add a is_correct column to indicate whether the prediction was correct or not:
plot_df <- plot_df %>% mutate(is_correct = class_pred == Purchase)
head(plot_df) Purchase LoyalCH PriceDiff class_pred is_correct
1 CH 0.500000 0.24 CH TRUE
2 CH 0.600000 -0.06 CH TRUE
3 CH 0.680000 0.40 CH TRUE
4 MM 0.400000 0.00 MM TRUE
5 CH 0.956535 0.00 CH TRUE
6 CH 0.965228 0.30 CH TRUERemember what we did in the Decision Tree example last week!
Simple confusion matrix:
confusion_matrix <-
table(expected=plot_df$Purchase, class_pred=plot_df$class_pred)
print(confusion_matrix)Nicer looking confusion matrix:
plot_confusion_matrix(as_tibble(confusion_matrix),
target_col = "expected",
prediction_col = "class_pred",
# Customizing the plot
add_normalized = TRUE,
add_col_percentages = FALSE,
add_row_percentages = FALSE,
counts_col = "n",
)🎯 ACTION POINT: How well does the model fit the data? What is your opinion?
Measure: Precision
Remember from Week 04 Lecture/Workshop (notebook can be found under📥W04 Lecture Files on Moodle):
PRECISION: Given all predictions for a specific class, how many were True Positives? In other words, Precision = True Positives/(True Positives + False Positives).
Let’s calculate the score for the CH class. That is, as if CH=“Yes” and MM=“No”.
# expected == CH & predicted == CH
total_correct_CH <- confusion_matrix["CH", "CH"]
# sum of samples predicted == CH
total_predicted_CH <- sum(confusion_matrix[, "CH"])
precision <- total_correct_CH/total_predicted_CH
cat(sprintf("%.2f%%", 100*precision))85.27%Measure: Recall
Also called True Positive Rate = True Positive/(True Positives + False Negatives)
# number of samples of brand CH
total_real_CH <- sum(confusion_matrix["CH", ])
recall <- total_correct_CH/total_real_CH
cat(sprintf("%.2f%%", 100*recall))87.75%Measure: F1-SCORE
F1-SCORE: A combination of Precision and Recall
\[ \operatorname{F1-score} = \frac{2 \times \operatorname{Precision} \times \operatorname{Recall}}{(\operatorname{Precision} + \operatorname{Recall})} \]
f1_score <- (2*precision*recall)/(precision + recall)
cat(f1_score)0.8649057Step 1.3: Visualize the SVM decision space
Here we will demonstrate how you could simulate some data to cover the entire feature space of the data we are modelling. What do we mean by that? By inspecting LoyalCH and PriceDiff, we see the range values these variables can assume:
filtered_data %>%
select(c(LoyalCH, PriceDiff)) %>%
summary() LoyalCH PriceDiff
Min. :0.000011 Min. :-0.6700
1st Qu.:0.325257 1st Qu.: 0.0000
Median :0.600000 Median : 0.2300
Mean :0.565782 Mean : 0.1465
3rd Qu.:0.850873 3rd Qu.: 0.3200
Max. :0.999947 Max. : 0.6400 💡 We can simulate data to account for all possible combinations of LoyalCH and PriceDiff. We achieve this using crossing, another tidyverse function:
- We feed
crossinga sequence of numbers that range from the minimal and maximal values of both variables, incremented by 0.1 values. - We then create a new variable
class_predwhich uses the SVM model object to predict the brand of orange juice purchased by the customer - Note that we say
newdata = .to indicate that we simply want to use the data set created withcrossingas our new data.
sim.data <-
crossing(LoyalCH = seq(0,1,0.05),
PriceDiff = seq(-1,1,0.1)) %>%
mutate(class_pred = predict(orange_svm_model, newdata = .))
head(sim.data)# A tibble: 6 × 3
LoyalCH PriceDiff class_pred
<dbl> <dbl> <fct>
1 0 -1 MM
2 0 -0.9 MM
3 0 -0.8 MM
4 0 -0.7 MM
5 0 -0.6 MM
6 0 -0.5 MM The data above is all synthetic (“fake”)! But it is very useful to colour the background of our plot.
We use geom_tile to show the area the SVM model identifies as Chinstrap penguins. We then use geom_point to overlay the actual data. Red dots in blue areas and vice versa indicate cases where the SVM model makes errors.
g <- (
plot_df %>%
ggplot()
# Tile the background of the plot with SVM predictions
+ geom_tile(data = sim.data, aes(x=LoyalCH, y=PriceDiff, fill = class_pred), alpha = 0.25)
# Actual data
+ geom_point(aes(x=LoyalCH, y=PriceDiff, colour = Purchase, shape = is_correct), size=2.5, stroke=0.95, alpha=0.7)
# Define X and Os
+ scale_shape_manual(values = c(4, 1))
# (OPTIONAL) Customizing the colours and theme of the plot
+ scale_x_continuous(labels=scales::percent)
+ scale_colour_startrek()
+ scale_fill_startrek()
+ theme_minimal()
+ theme(panel.grid = element_blank(), legend.position = 'bottom', plot.title = element_text(hjust = 0.5))
+ labs(x = 'Customer brand loyalty for CH', y = 'Sale price of MM less sale price of CH', fill = 'Brand', colour = 'Brand', shape = 'Correct prediction?', title = sprintf('Overall Training Accuracy = %.2f %%', 100*(sum(plot_df$correct)/nrow(plot_df))))
)
g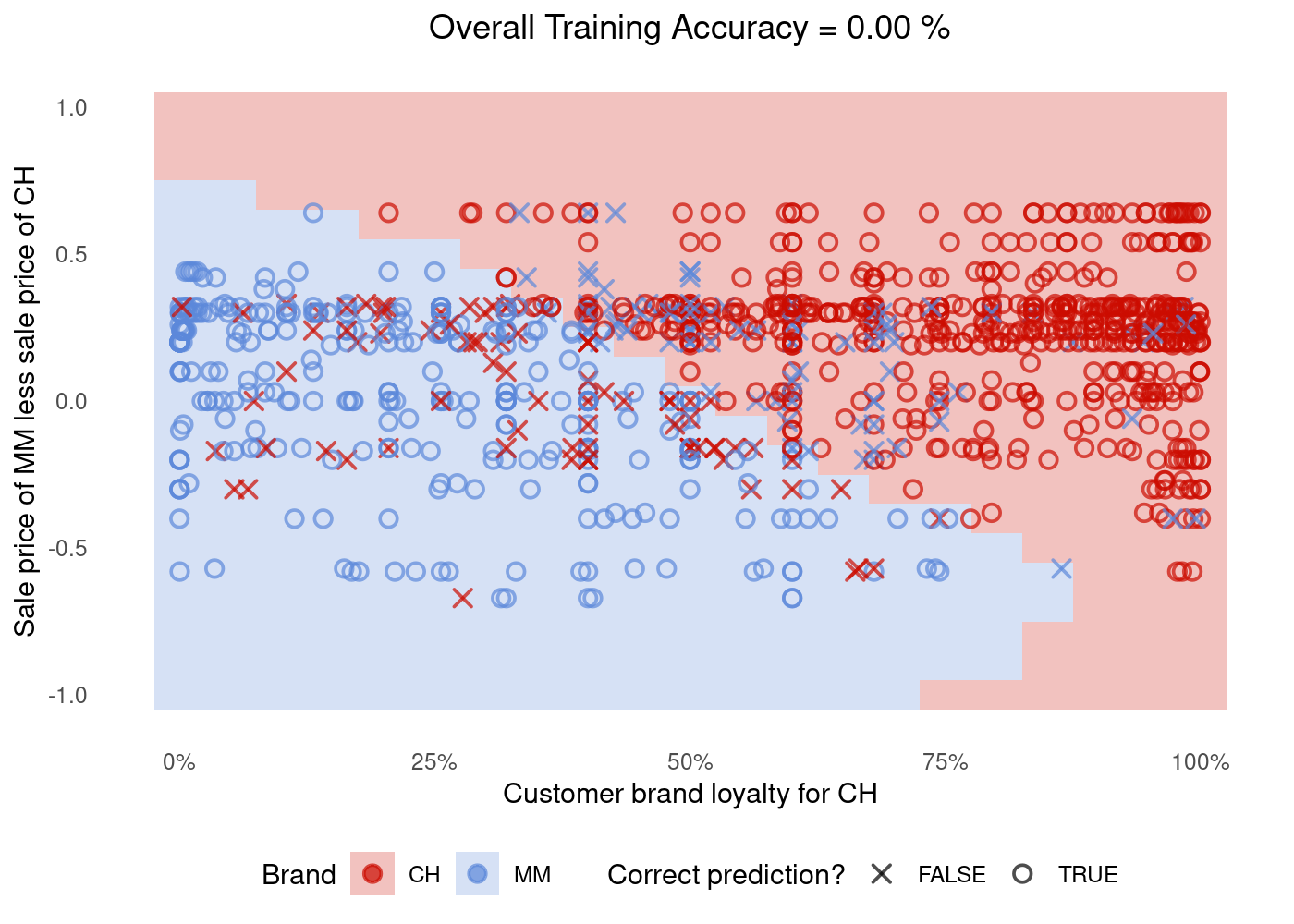
🤝 WORKING TOGETHER In pairs, discuss what you see in the plot:
- What do the shape of dots represent? The
XandOs?
your text here
- What do the colours of the dots represent?
your text here
- What do the background colour in the plot represent?
your text here
- Can you point in the plot roughly which dots you would expect to be the support vectors?
Step 2: Doing the same with tidymodels (20 min)
Step 2: Doing the same with tidymodels (20 min)
The function svm from library(e1071) package is not the only way to run SVM in R. The package parsnip also have its own SVM functions. The functionality is roughly the same but there are differences in how you write the code.
parsnip already comes installed in tidymodels, so we do not need to import or install anything else.
Step 2.1 Training the SVM model
We specify a radial basis function SVM (see the part about kernels in the 🗓️ Week 05 lecture) with the function svm_rbf.
In the spirit of tidyverse, we pipe the SVM algorithm into the fit function, where we can define the R formula like we have been doing with other algorithms:
orange_tidymodel <-
svm_rbf() %>%
set_mode('classification') %>%
fit(Purchase ~ ., data = filtered_data)
orange_tidymodelparsnip model object
Support Vector Machine object of class "ksvm"
SV type: C-svc (classification)
parameter : cost C = 1
Gaussian Radial Basis kernel function.
Hyperparameter : sigma = 1.65374574360967
Number of Support Vectors : 432
Objective Function Value : -386.8495
Training error : 0.170093
Probability model included. 🎯 ACTION POINT: Compare the output above to that of another colleague. Why don’t you get the exact same output?
your personal notes go here
💡 If you want to try different kernels, you will need to replace svm_rbf() by svm_linear() or svm_poly().
Step 2.2: Goodness-of-Fit of the SVM
Let’s investigate how well our model fits the data. Let’s reuse the model we trained (orange_tidymodel) and predict the same samples we used to train it.
Function augment(<model>, <df>) of tidymodels applies a model to a dataframe and return the same data plus a few columns:
plot_df <- augment(orange_tidymodel, filtered_data)
head(plot_df)🎯 ACTION POINT: How is the plot_df data frame above different to the first plot_df we created in Step 1.2?
your notes go here
Add a is_correct column to indicate whether the prediction was correct or not:
plot_df <- plot_df %>% mutate(is_correct = .pred_class == Purchase)
head(plot_df)# A tibble: 6 × 7
Purchase LoyalCH PriceDiff .pred_class .pred_CH .pred_MM is_correct
<fct> <dbl> <dbl> <fct> <dbl> <dbl> <lgl>
1 CH 0.5 0.24 CH 0.862 0.138 TRUE
2 CH 0.6 -0.06 CH 0.718 0.282 TRUE
3 CH 0.68 0.4 CH 0.876 0.124 TRUE
4 MM 0.4 0 MM 0.174 0.826 TRUE
5 CH 0.957 0 CH 0.879 0.121 TRUE
6 CH 0.965 0.3 CH 0.876 0.124 TRUE Measure: Precision
You don’t need to calculate precision by hand, just use the precision() function from tidymodels:
plot_df %>% precision(Purchase, .pred_class) %>% head()# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 precision binary 0.856Measure: Recall
You don’t need to calculate recall by hand, just use the recall() function from tidymodels:
plot_df %>% recall(Purchase, .pred_class) %>% head()# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 recall binary 0.867Measure: F1-score
You don’t need to calculate F1-score by hand, just use the f_meas() function from tidymodels:
plot_df %>% f_meas(Purchase, .pred_class) %>% head()# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 f_meas binary 0.861(Optional) ROC curve
Plot the ROC curve for class Purchase=="CH" :
plot_df %>%
roc_curve(Purchase, .pred_CH) %>%
autoplotWarning: Returning more (or less) than 1 row per `summarise()` group was deprecated in
dplyr 1.1.0.
ℹ Please use `reframe()` instead.
ℹ When switching from `summarise()` to `reframe()`, remember that `reframe()`
always returns an ungrouped data frame and adjust accordingly.
ℹ The deprecated feature was likely used in the yardstick package.
Please report the issue at <https://github.com/tidymodels/yardstick/issues>.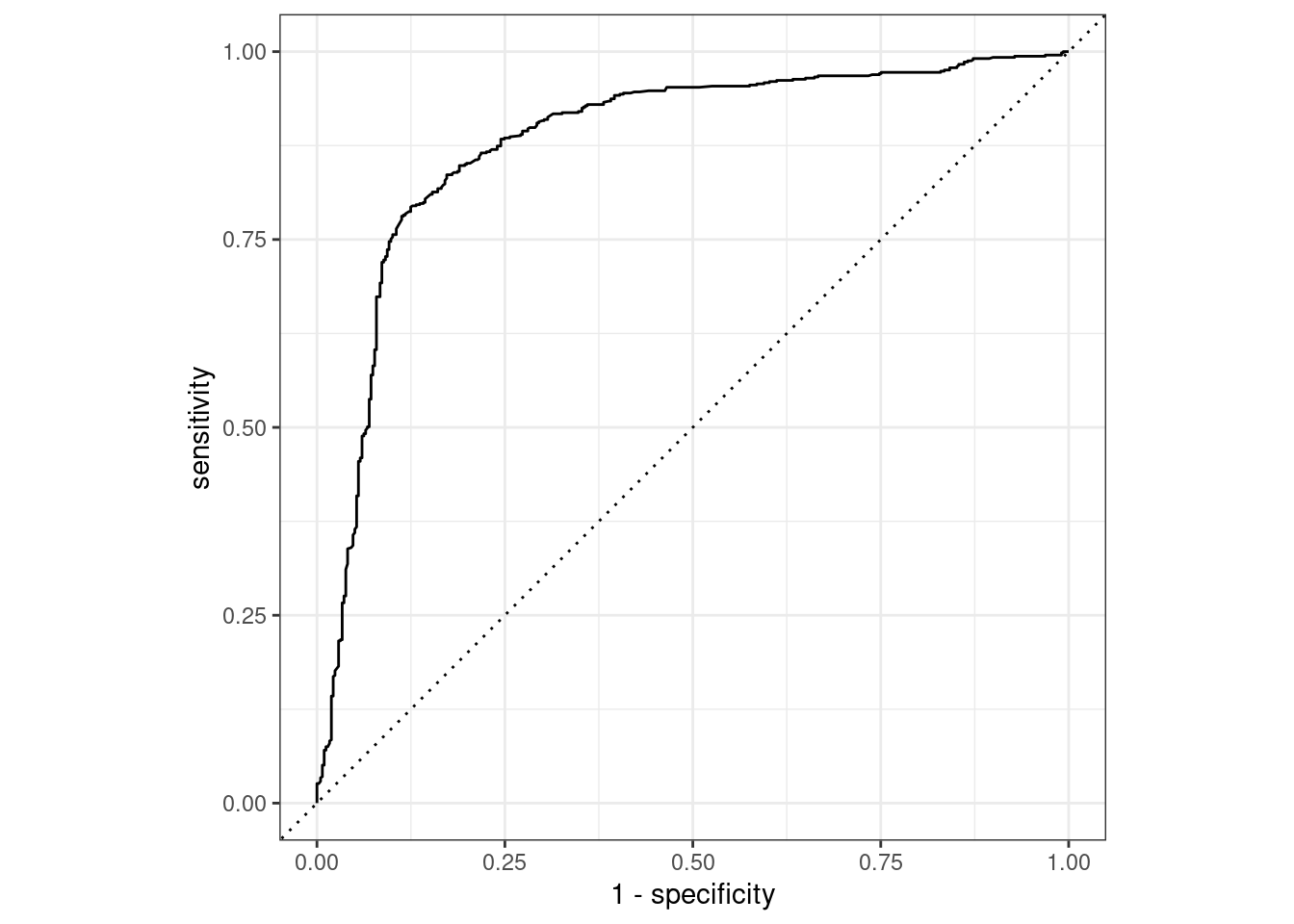
🏠 Take-home exercise Q1:
Edit the cell below modifying event_level from "second" to "first". Why do you get different results? What do you think is going on?
plot_df %>% f_meas(Purchase, .pred_class, event_level=...)💡Tip: Read the documentation of f_meas to understand what event_level represents. (Type ?f_meas)
💡 Gold Tip: note the Levels of the factor variable called Purchase:
plot_df$Purchase🏠 Take-home exercise Q2:
Create a plot of the confusion matrix for the orange_tidymodel like we did in Step 1.
## your code goes here🏠 Take-home exercise Q3:
Create a plot of SVM decision space for the orange_tidymodel like we did in Step 1.
## your code goes here🎯 ACTION POINT: If you were to run the SVM algorithm by yourself in another dataset, which version would you prefer, the one in Step 1 or the one in Step 2?
your notes go here
Step 3: What about regression? (30 min)
Step 3: What about regression? (30 min)
Here we will be using the 📈 SMarket data.
Step 3.1: Train the model
Let’s select just the predictors Volume and Lag1 and fit a regression model to predict Today:
# Remove Direction, otherwise we would be "cheating"
filtered_data <- ISLR2::Smarket %>% select(Today, Volume, Lag1)
smarket_tidymodel <-
svm_rbf() %>%
set_mode('regression') %>%
fit(Today ~ ., data = filtered_data)
smarket_tidymodelparsnip model object
Support Vector Machine object of class "ksvm"
SV type: eps-svr (regression)
parameter : epsilon = 0.1 cost C = 1
Gaussian Radial Basis kernel function.
Hyperparameter : sigma = 1.79706099888903
Number of Support Vectors : 1117
Objective Function Value : -760.8719
Training error : 0.927028 Step 3.2: Goodness-of-Fit of the SVM
Since the target variable is continuous, not discrete, we cannot plot confusion matrix nor anything like that. We will have to go back to the idea of residuals (🗓️ Week 02 Lecture & 💻 Week 03 - Lab).
plot_df <-
augment(smarket_tidymodel, filtered_data) %>%
mutate(row_number=row_number()) # adding this here just to make our plot easier
plot_df %>% head()# A tibble: 6 × 6
Today Volume Lag1 .pred .resid row_number
<dbl> <dbl> <dbl> <dbl> <dbl> <int>
1 0.959 1.19 0.381 -0.0565 1.02 1
2 1.03 1.30 0.959 -0.256 1.29 2
3 -0.623 1.41 1.03 -0.263 -0.360 3
4 0.614 1.28 -0.623 0.261 0.353 4
5 0.213 1.21 0.614 -0.153 0.366 5
6 1.39 1.35 0.213 -0.149 1.54 6🎯 ACTION POINT: What do the different columns mean?
your text go here
Now, let’s look at the distribution of residuals and let’s mark the absolute residuals above 2 to flag the worst predictions (2 was an arbitrary choice, it all depends on the context):
g <- (
ggplot(plot_df, aes(x=row_number, y=.resid))
+ geom_point(alpha=0.6)
+ theme_bw()
+ geom_hline(yintercept = c(-2,2), color="red", linetype="dashed")
+ labs(title="Distribution of residuals (the closer to zero the better)")
)
g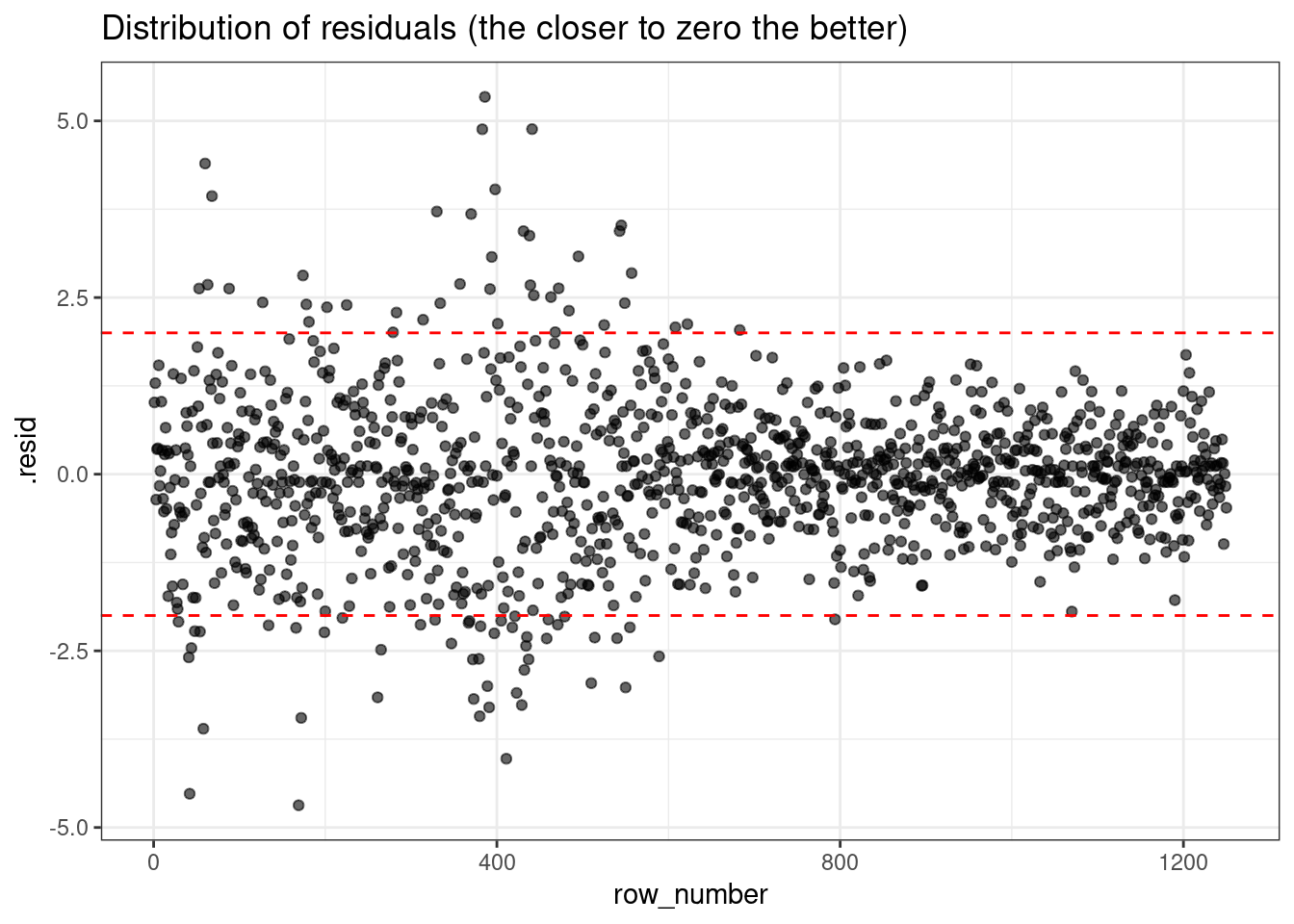
Measure: Mean Absolute Error (MAE)
\[ MAE = \frac{\sum_{i=1}^{n}{|y_i - \hat{y}_i|}}{n} \]
plot_df %>% mae(Today, .pred)# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 mae standard 0.789Measure: Root Mean Squared Error (RMSE)
\[ RMSE = \frac{\sum_{i=1}^{n}{(y_i - \hat{y}_i)^2}}{n} \]
plot_df %>% rmse(Today, .pred)# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 rmse standard 1.09🎯 ACTION POINT: Would a better model have a larger or smaller value of MAE/RMSE?
your text goes here
Step 3.3 Visualize the SVM decision space (Regression)
Now, let’s replicate what we did in Step 1.3 only this time for the Smarket data and using predictions from the smarket_tidymodel.
We start by summarising the data. We want to find out the minimum and maximal values that the columns Volumn and Lag1 reach:
filtered_data %>%
select(c(Volume, Lag1)) %>%
summary() Volume Lag1
Min. :0.3561 Min. :-4.922000
1st Qu.:1.2574 1st Qu.:-0.639500
Median :1.4229 Median : 0.039000
Mean :1.4783 Mean : 0.003834
3rd Qu.:1.6417 3rd Qu.: 0.596750
Max. :3.1525 Max. : 5.733000 Then, we create a simulated dataset with combinations of the Volume and Lag1 columns:
sim.data <-
crossing(Volume = seq(0,3.5,0.1),
Lag1 = seq(-5,6,0.2))
sim.data <- augment(smarket_tidymodel, sim.data)
head(sim.data)# A tibble: 6 × 3
Volume Lag1 .pred
<dbl> <dbl> <dbl>
1 0 -5 -0.0103
2 0 -4.8 -0.0103
3 0 -4.6 -0.0103
4 0 -4.4 -0.0103
5 0 -4.2 -0.0103
6 0 -4 -0.0103💡 Look at the entire dataset with View(sim.data)
Looking back at the plot of residuals, let’s flag the worst predictions, those with an absolute residual above 2.
plot_df$residual_above_2 <- (plot_df$.resid) > 2g <- (
plot_df %>%
ggplot()
## Tile the background of the plot with SVM predictions
+ geom_tile(data = sim.data, aes(x=Volume, y=Lag1, fill = .pred), alpha = 0.45)
## Actual data
+ geom_point(aes(x=Volume, y=Lag1, colour = residual_above_2, shape = residual_above_2, alpha=residual_above_2), size=2.5, stroke=0.95)
## Define X and Os
+ scale_shape_manual(values = c(4, 1))
+ scale_fill_viridis_c()
+ scale_color_manual(values=c("black", "red"))
+ scale_alpha_manual(values=c(0.1, 0.7))
## (OPTIONAL) Customizing the colours and theme of the plot
+ theme_minimal()
+ theme(panel.grid = element_blank(), legend.position = 'bottom', plot.title = element_text(hjust = 0.5))
+ labs(x = 'Volume', y = 'Lag 1', fill = "Today's Prediction", colour = 'Residual above 2?', shape = 'Residual above 2?', alpha='Residual above 2?', title='Worst predictions are marked as red circles')
)
g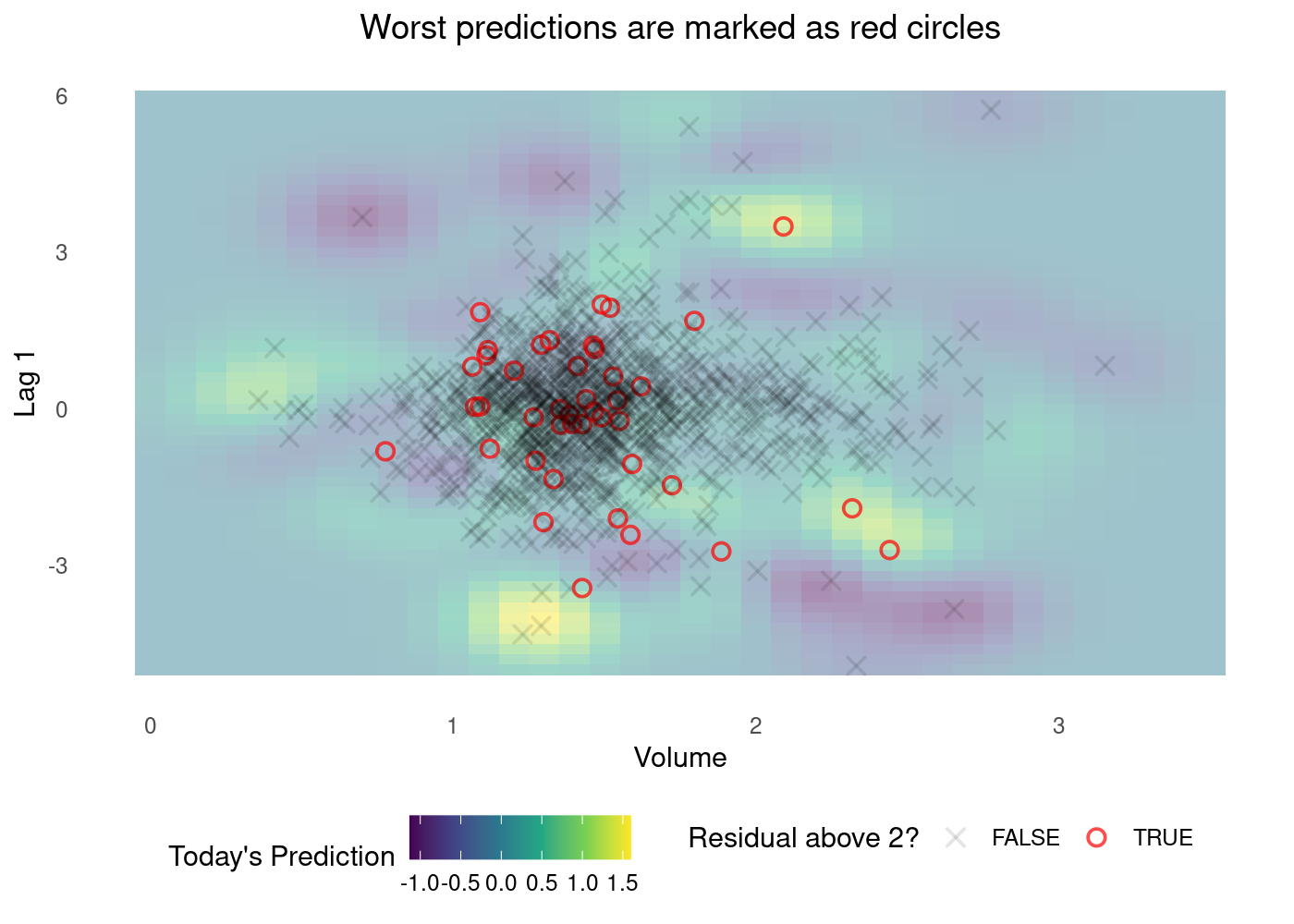
💡 The plot above might not be as easy to understand as the one for classification.
Step 3.3: Understand the parameters of svm_rbf
The svm_rbf function has three parameters you can tune:
costrbf_sigmamargin
🎯 ACTION POINT: Train your abilities to interact with code documentation. Type ?svm_rbf and hit ENTER. What do these parameters represent?
💡 Tip: At the bottom of the help page, you will find a link to kernlab engine details that has more useful info about SVM RBF.
your text goes here
Step 3.4: Tweak the parameters
🤝 WORKING TOGETHER In pairs, change the values of cost, rbf_sigma and margin in the chunk below and run the other two chunks of code to look at the distribution of residuals and summary metrics.
Discuss your findings. Can you find any combination of values that makes the model better? Or any that makes it worse?
alternative_smarket_model <-
svm_rbf(cost=1, rbf_sigma=10, margin=0.9) %>%
set_mode('regression') %>%
fit(Today ~ ., data = filtered_data)
alternative_smarket_modelparsnip model object
Support Vector Machine object of class "ksvm"
SV type: eps-svr (regression)
parameter : epsilon = 0.9 cost C = 1
Gaussian Radial Basis kernel function.
Hyperparameter : sigma = 10
Number of Support Vectors : 401
Objective Function Value : -195.7568
Training error : 0.832623 Residuals plot
plot_df <-
augment(alternative_smarket_model, filtered_data) %>%
mutate(row_number=row_number()) ## adding this here just to make our plot easier
g <- (
ggplot(plot_df, aes(x=row_number, y=.resid))
+ geom_point(alpha=0.6)
+ theme_bw()
+ geom_hline(yintercept = c(-2,2), color="red")
)
g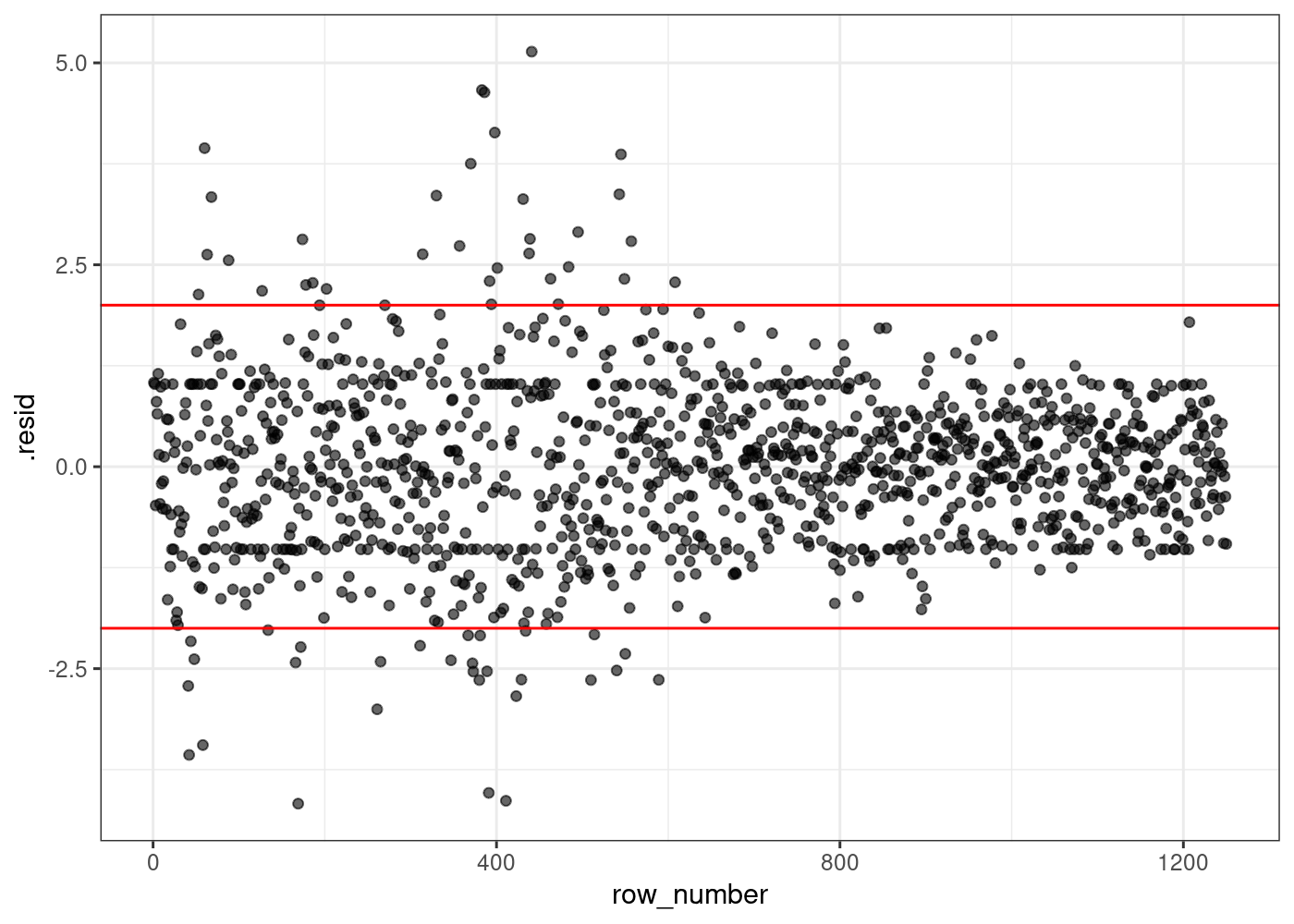
Metrics
## Use the vectorized version of MAE and RMSE functions
plot_df %>% summarise(mae=mae_vec(Today, .pred),
rmse=rmse_vec(Today, .pred))# A tibble: 1 × 2
mae rmse
<dbl> <dbl>
1 0.787 1.04🏠 Take-home exercise Q4:
- Retrain the model in Step 3 for
Smarket, this time usingsvm_linearinstead ofsvm_rbf. - Reuse the code in Step 3.4 to replicate the plots and metric calculations for this new model.
- Which model fits the target variable (
Today) better?
# your code goes hereyour text goes here
Want to take it to the next level? (Optional)
Want to take it to the next level? (Optional)
In a separate, more advanced, bonus lab roadmap we show how to perform k-fold cross-validation using tidymodels to tune the parameters of SVM automatically.
It is optional, you don’t need to read it, but it might be the best way to solidify your knowledge of SVM and its parameters.
