🛣️ LSE DS202A 2025: Week 05 - Lab Roadmap
🎯 Learning Outcomes
By the end of this lab, you will be able to:
Implement k-nearest neighbors with hyperparameter tuning
- Build k-NN models using
tidymodels, understand how different k values affect decision boundaries, and use cross-validation to systematically optimize the number of neighbors for best performance.
- Build k-NN models using
Apply advanced cross-validation techniques for model evaluation
- Implement grouped cross-validation for clustered data structures, understand when to use different CV strategies, and interpret cross-validation results to select optimal hyperparameters.
Build and compare tree-based models
- Construct decision trees, random forests, and XGBoost models, visualize decision tree structures, and understand the trade-offs between interpretability and predictive performance across different ensemble methods.
Master hyperparameter tuning workflows
- Use
tidymodels’ tune_grid() framework to systematically test different parameter combinations, create recipes for data preprocessing, and build complete workflows that combine model specifications with feature engineering steps.
- Use
📚 Preparation
Downloading the data sets
Download the lab’s .qmd notebook
Click on the link below to download the .qmd file for this lab. Save it in the DS202A folder you created in the first week. If you need a refresher on the setup, refer back to Part II of W01’s lab.
Import libraries
library("rpart.plot")
library("tidymodels")
library("tidyverse")
library("viridis")
library("rsample")
library("doParallel")
library("ranger")
library("xgboost")Part I - k-Nearest Neighbors and Cross-Validation
Understanding decision boundaries in 2D feature space
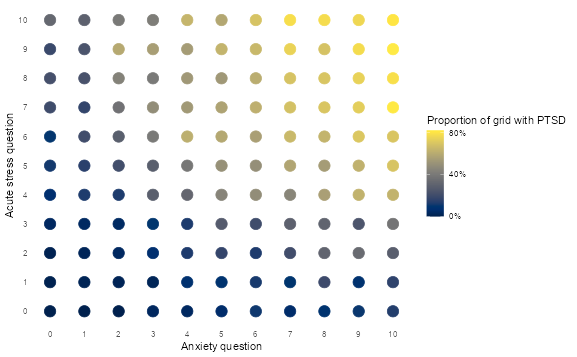
👨🏻🏫 TEACHING MOMENT: Your instructor will demonstrate how different machine learning algorithms create different decision boundaries in a 2D feature space using the PTSD data.
ptsd <-
read_csv("data/ptsd-for-knn.csv") |>
mutate(ptsd = as.factor(ptsd))
# Explore the feature space
ptsd |>
group_by(anxiety, acute_stress) |>
summarise(ptsd = mean(ptsd == 1, na.rm = TRUE)) |>
ungroup() |>
ggplot(aes(anxiety, acute_stress, colour = ptsd)) +
geom_point(size = 5) +
scale_x_continuous(breaks = 0:10) +
scale_y_continuous(breaks = 0:10) +
scale_colour_viridis(
labels = percent,
breaks = c(0, 0.4, 0.8),
option = "cividis"
) +
theme_minimal() +
theme(panel.grid = element_blank()) +
labs(
x = "Anxiety question",
y = "Acute stress question",
colour = "Proportion of grid with PTSD"
)
Challenge: Build a cross-validated k-NN model
Your mission: Create a k-NN model that uses 10-fold cross-validation to find the optimal number of neighbors for predicting PTSD.
Function glossary for this section:
initial_split()- split data into train/testvfold_cv()- create cross-validation folds
nearest_neighbor()- specify k-NN modeltune()- mark parameters for tuningrecipe()- data preprocessing stepsstep_normalize()- standardize featuresworkflow()- combine model + recipetune_grid()- perform cross-validationcollect_metrics()- extract CV resultsselect_best()- choose optimal parameters
Helpful resources:
Your tasks:
- Split the data (75% train, 25% test, stratified by ptsd)
- Create 10-fold CV on training data
- Set up a k-NN model with
neighbors = tune() - Create a recipe that normalizes features
- Build a workflow combining model + recipe
- Test different k values:
c(1, 5, 10, 20, 50, 100, 200) - Visualize how performance changes with k
- Select the best k and evaluate on test set
# Code hereDiscussion questions:
- Which k value performed best and why?
- What happens with very small vs very large k values?
- How do the cross-validation results compare to test set performance?
Part II - Tree-Based Models and Ensemble Methods
Understanding tree growth and ensembles
👨🏻🏫 TEACHING MOMENT: Your instructor will demonstrate:
- How a basic decision tree splits data
- How trees can grow and overfit
- Introduction to ensemble methods (bagging, random forests)
- How XGBoost incorporates learning from mistakes
Challenge: Compare tree-based models
Your mission: Build and compare three tree-based models for predicting childcare costs.
Function glossary for this section:
initial_split()- split data into train/testsliding_period()- time-series CV with temporal ordering (⚠️requires the years to be converted from double to dates!)decision_tree()- single decision treerand_forest()- random forest modelboost_tree()- XGBoost modelrpart.plot()- visualize decision treeupdate_role()- assign variable roles in recipe
# ------------------------------------------------------------------------------
# 1. Load and prepare data
# ------------------------------------------------------------------------------
childcare <- read_csv("data/childcare-costs.csv")
# Split by time:
# - Train: 2008–2016
# - Test: 2017–2018
# We sort by year to preserve temporal order
# and create a Date column (`study_date`) for time-series cross-validation.
childcare_train <-
childcare %>%
filter(study_year <= 2016) %>%
arrange(study_year) %>%
mutate(study_date = as.Date(paste0(study_year, "-01-01")))
childcare_test <-
childcare %>%
filter(study_year > 2016) %>%
mutate(study_date = as.Date(paste0(study_year, "-01-01")))
# -------------------------------------------------------# ------------------------------------------------------------------------------
# 2. Create time-series cross-validation folds
# ------------------------------------------------------------------------------
# We use sliding_period() instead of vfold_cv()
# to ensure no data leakage from future to past.
# The window expands each year, always predicting the next one.
childcare_cv <- sliding_period(
childcare_train,
index = study_date,
period = "year",
lookback = Inf, # Expanding window: train on all prior years
assess_stop = 1, # Test on 1 future year
step = 1 # Move forward 1 year each time
)
# This yields 8 folds (2008–2016)
length(childcare_cv$splits)The approach above is appropriate when you want to predict future costs in counties you already have data for. However, if your goal is to predict costs in entirely new counties (e.g., expanding to new geographic markets), you need to combine time-series CV with county grouping:
library(tidyverse)
library(rsample)
# --- 1. Sample holdout counties once (never used in training) ----------
set.seed(123)
holdout_counties <- childcare_train %>%
distinct(county_name) %>%
slice_sample(prop = 0.2) %>%
pull(county_name)
# --- 2. Build expanding-window time folds with fixed holdout counties --
val_years <- 2013:2016
childcare_cv_tbl <- tibble(val_year = val_years) %>%
mutate(
splits = map(val_year, \(year) {
make_splits(
list(
# Train: all years before validation year, excluding holdout counties
analysis = which(
childcare_train$study_year < year &
!(childcare_train$county_name %in% holdout_counties)
),
# Validate: current year, only holdout counties
assessment = which(
childcare_train$study_year == year &
childcare_train$county_name %in% holdout_counties
)
),
data = childcare_train
)
}),
id = glue::glue("Year_{val_year}")
)
# --- 3. Convert to a valid rset object (tidymodels-compatible) ----------
childcare_cv <- new_rset(
splits = childcare_cv_tbl$splits,
ids = childcare_cv_tbl$id,
attrib = tibble(val_year = childcare_cv_tbl$val_year),
subclass = c("manual_rset", "rset")
)
childcare_cvWhat this creates:
- Fold 1: Train on 2008-2012 (80% of counties) → Validate on 2013 (20% holdout counties)
- Fold 2: Train on 2008-2013 (80% of counties) → Validate on 2014 (20% holdout counties)
- Fold 3: Train on 2008-2014 (80% of counties) → Validate on 2015 (20% holdout counties)
- Fold 4: Train on 2008-2015 (80% of counties) → Validate on 2016 (20% holdout counties)
This ensures:
- No time leakage (always train on past, validate on future)
- No county leakage (holdout counties never appear in training)
- True generalization to new geographic areas testing
- Expanding window mimics realistic deployment (more training data over time)
Now that we’ve loaded the data and set the cross-validation, let’s write a pre-processing recipe:
# ------------------------------------------------------------------------------
# 3. Recipe: preprocessing pipeline
# ------------------------------------------------------------------------------
# We specify our modeling formula and tell tidymodels which variables
# should NOT be used as predictors (like county/state identifiers or study_date/study_year).
childcare_rec <-
recipe(mcsa ~ ., data = childcare_train) %>%
update_role(starts_with(c("county", "state")), new_role = "id") %>%
update_role(c("study_date","study_year"), new_role = "id") # used only for CV indexing, not modelingTask 1: Decision Tree
Build a simple decision tree and visualize it:
# Code hereClass discussion: What does the tree visualization show? Which variables are most important?
Task 2: Random Forest
Build a random forest with mtry = round(sqrt(52), 0) and trees = 1000:
# Code hereTask 3: XGBoost with hyperparameter tuning
Build an XGBoost model and tune the learning rate using these values: c(0.0001, 0.001, 0.01, 0.1, 0.2, 0.3):
# Code hereTask 4: Compare all models
Create a visualization comparing the RMSE of all three models on the test set.
# Code hereFinal discussion:
- Which model performed best and why?
- What are the trade-offs between interpretability and performance?
- When might you choose each type of model?
