🗓️ Week 05:
Support Vector Machines (SVM)
Non-linear algorithms
10/28/22
Support Vector Machines
Support Vector Machines for Classification
- Considered one of the best out-of-the-box classifiers (ISLR)
- Able to accommodate non-linear decision boundaries
Building Intuition: the Hyperplane
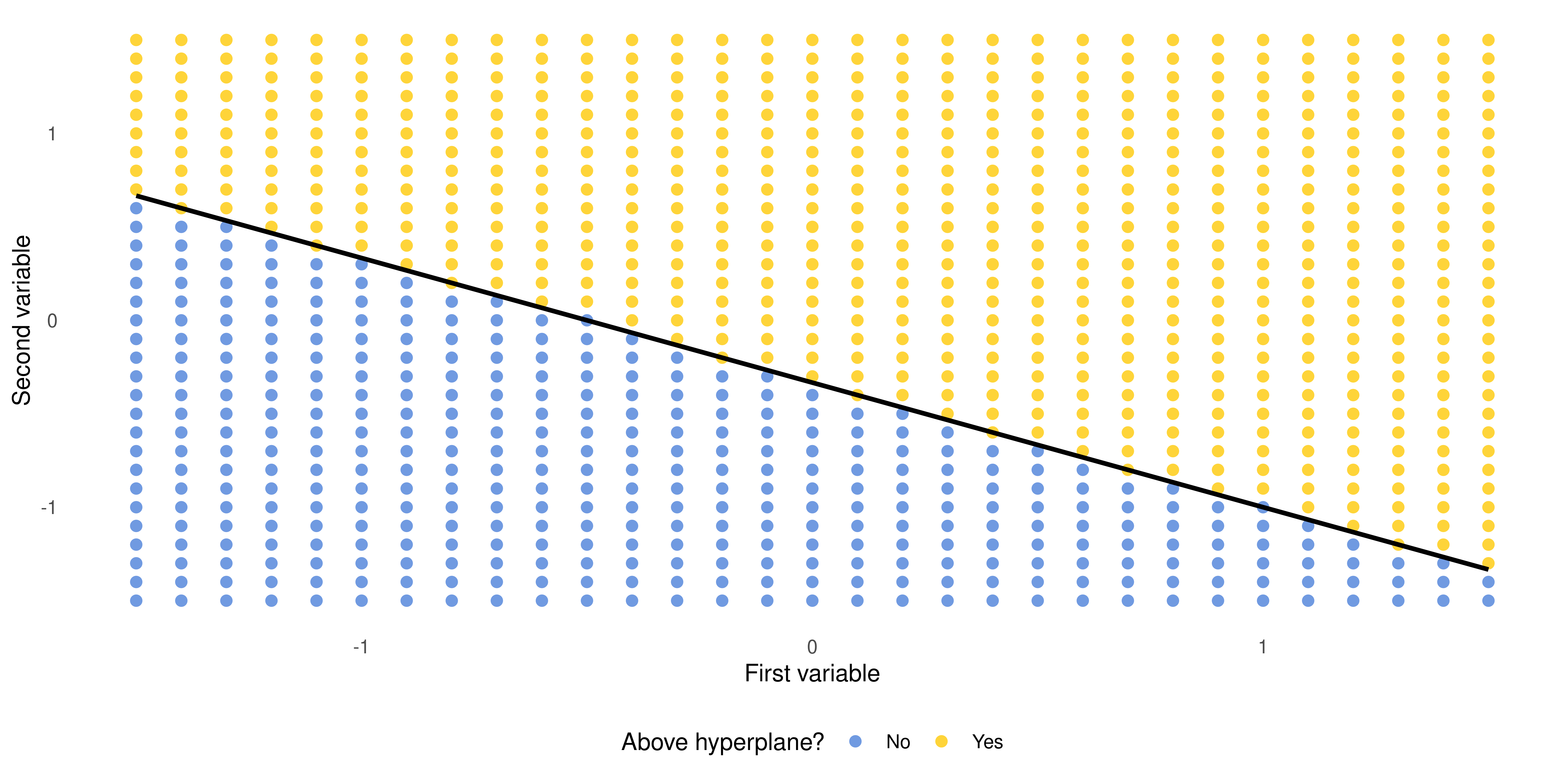
Building Intuition: the Hyperplane (cont.)
When \(1 + 2x_1 + 3x_2 < 0\)
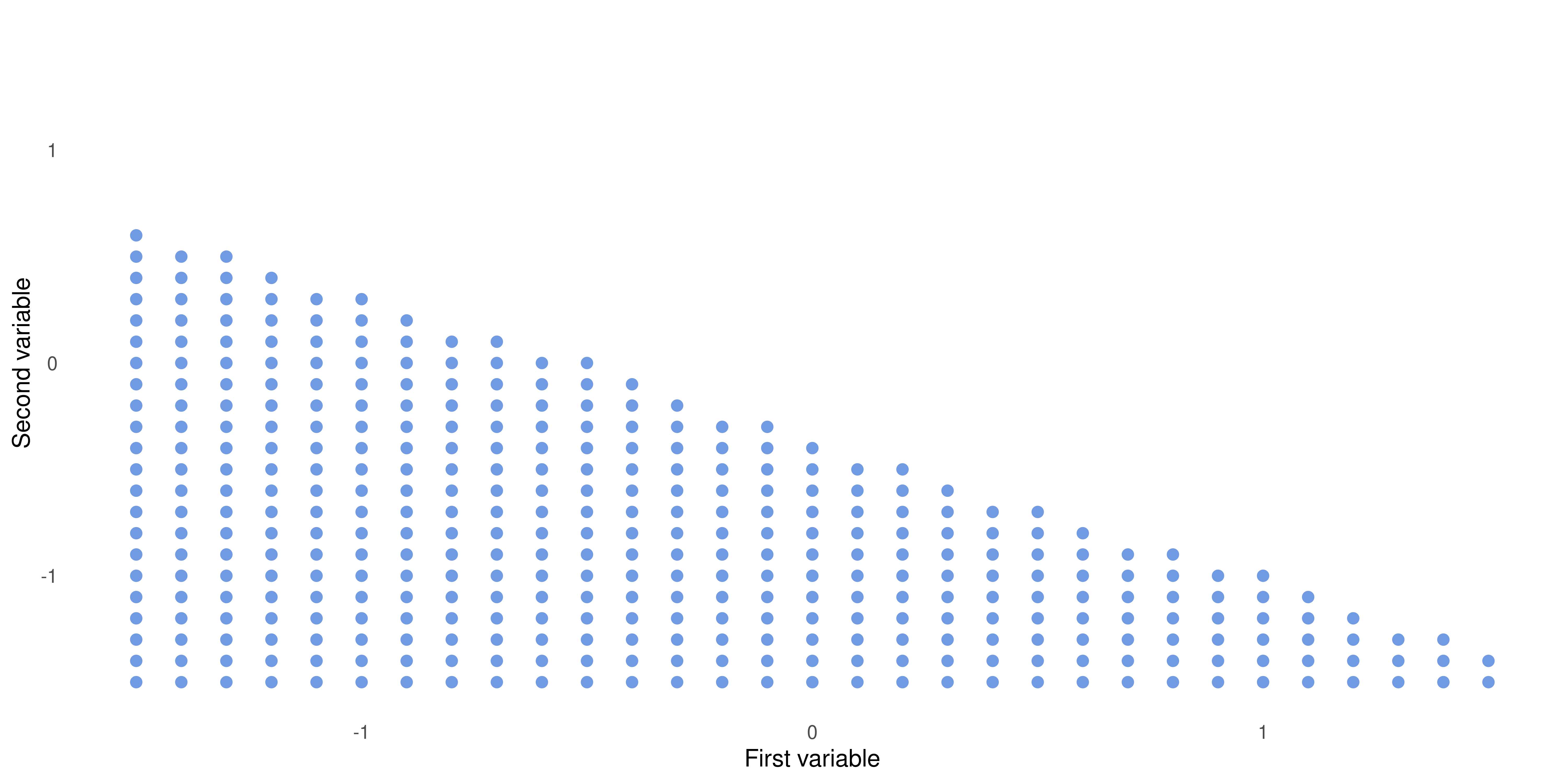
Building Intuition: the Hyperplane (cont.)
When \(1 + 2x_1 + 3x_2 > 0\)
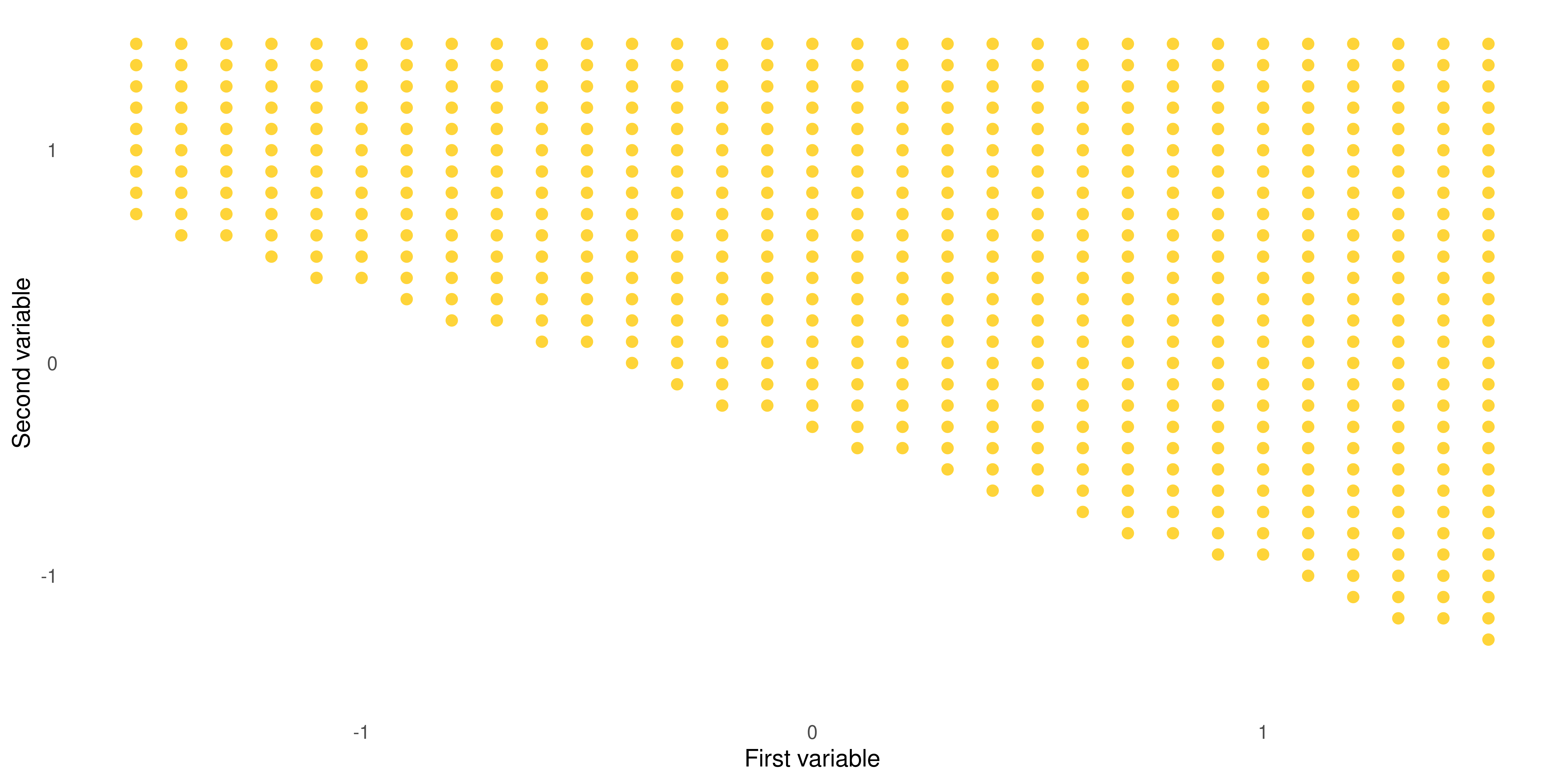
Building Intuition: the Hyperplane (cont.)
When \(1 + 2x_1 + 3x_2 = 0\)

The Maximal Marginal Classifier
The linearly separable case:
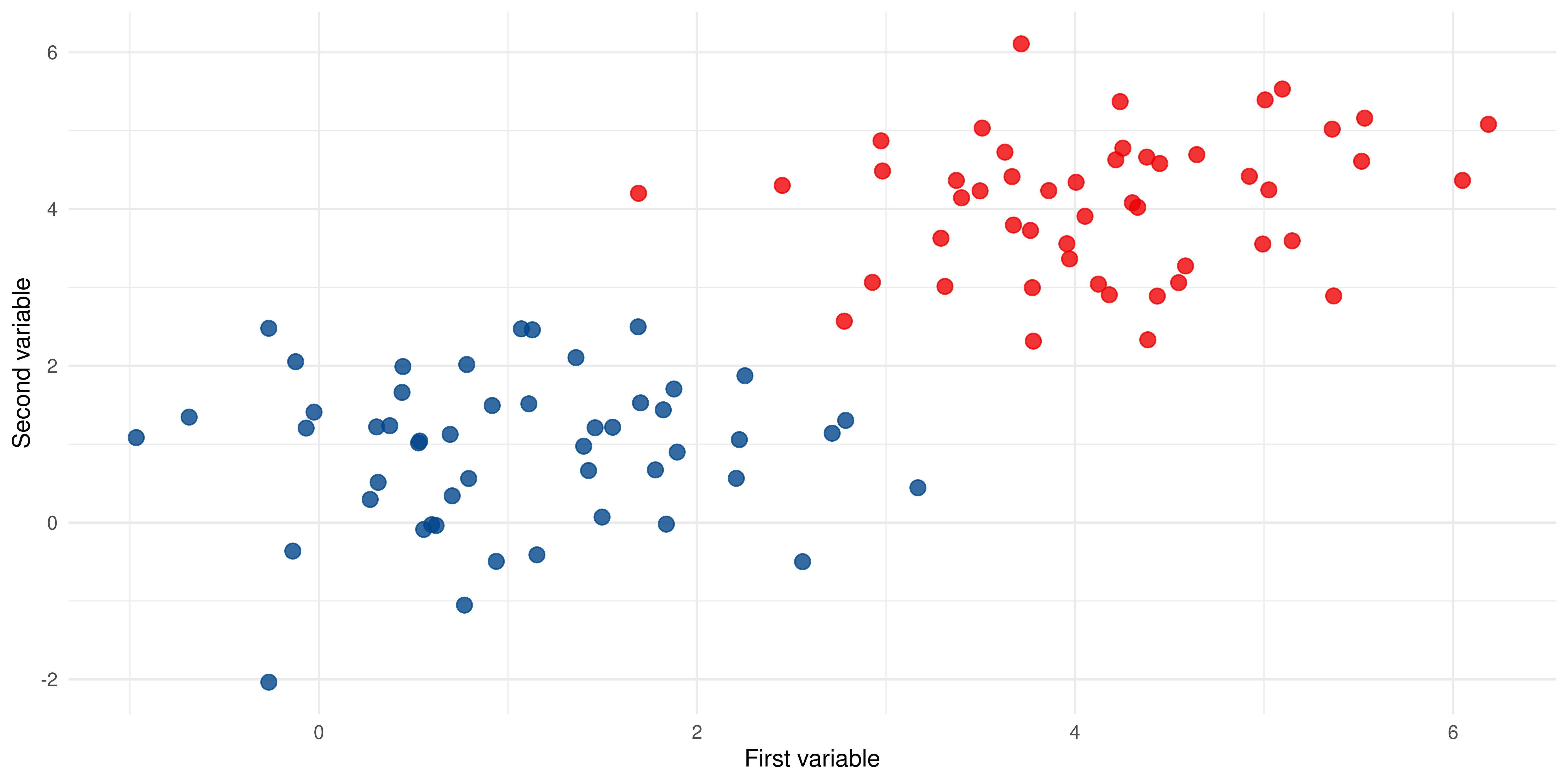
Identifying support vectors
The SV’s represent the so-called support vectors:

When data is not linearly separable
Suppose we have a case that is not linearly separable like this. We have two classes but class 1 is “sandwiched” in between class 2.
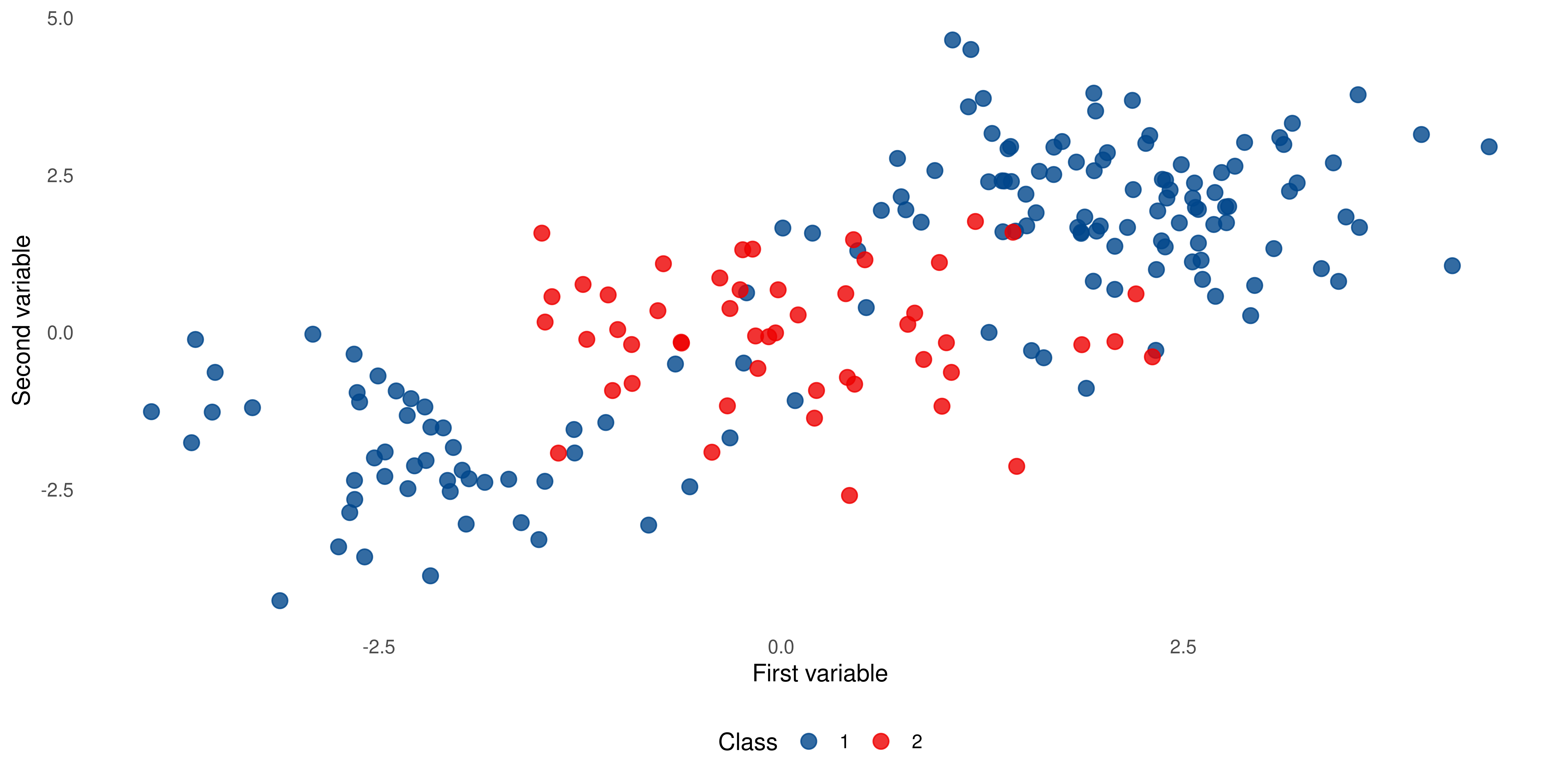
Enter support vector machines
Let’s start with a (linear) support vector classifier function
\[ f(x) = \beta_0 + \sum_{i\in\mathcal{S}}^{} \alpha_i\left\langle x_i, x_{i'} \right\rangle \]
A couple of points:
- \(\left\langle x_i, x_{i'} \right\rangle\) is the inner product 1 of two observations
- \(\alpha_i\) is a parameter fitted for all pairs of training observations
- \(i\in\mathcal{S}\) indicates observations that are support vectors (all other observations are ignored by setting all \(\alpha_i \notin \mathcal{S}\) to zero.)
Enter support vector machines (cont.)
- We can replace \(\left\langle x_i, x_{i'} \right\rangle\) with a of the form \(K(x_i, x_{i'})\) where \(K\) is a kernel.
Two well-known kernels:
- Polynomial \(K(x_i, x_{i'}) = (1 + \sum_{j = 1}^{p} x_{ij}x_{i'j})^d\) where \(d > 1\).
- Radial or “Gaussian” \(K(x_i, x_{i'}) = \text{exp}(-\gamma\sum_{j=1}^{p}(x_{ij}-x_{i'j})^2)\) where \(\gamma\) is a positive constant.
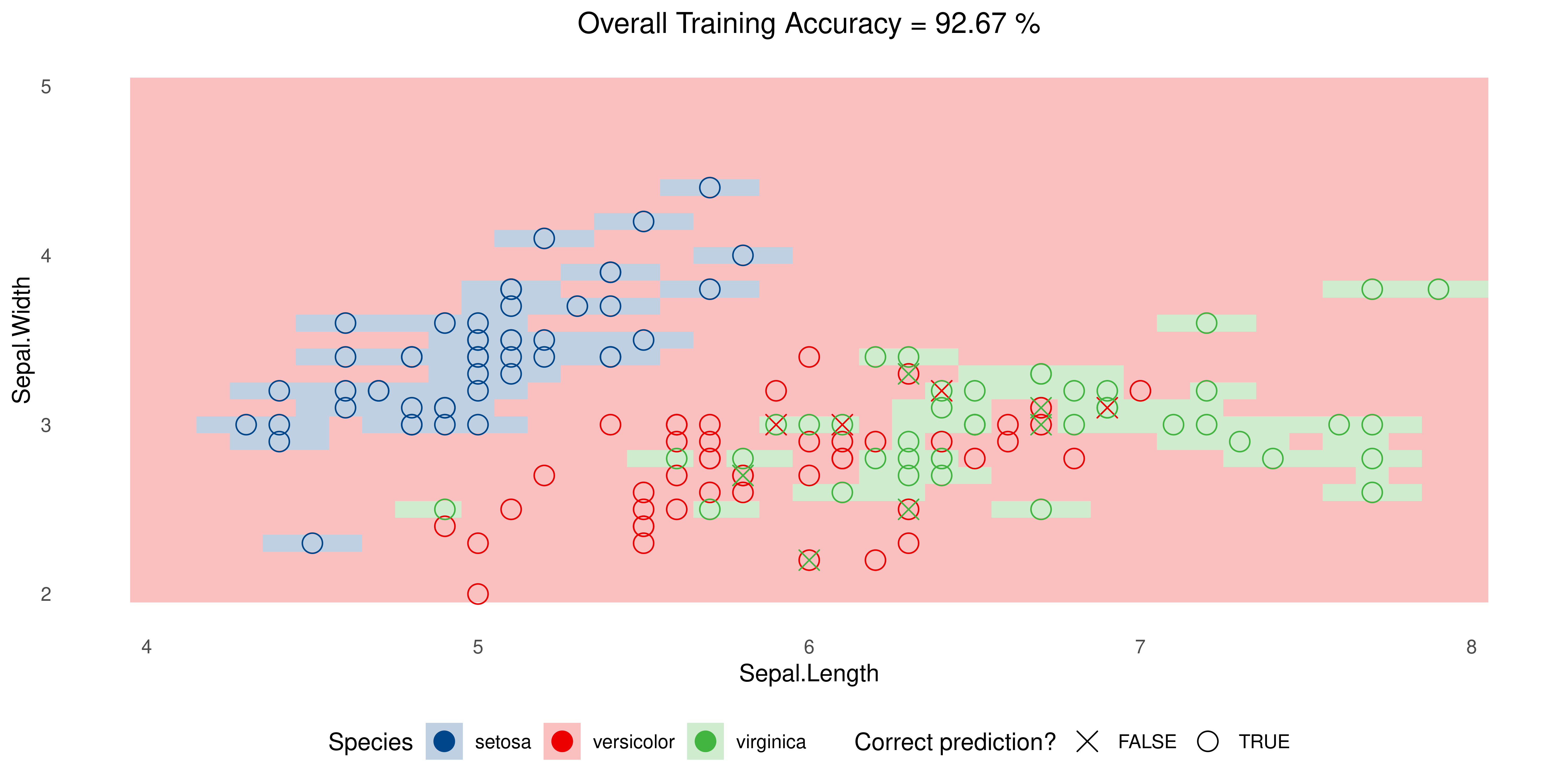
Example: Iris data
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosaSVM with Linear Kernel

SVM with Polynomial Kernel
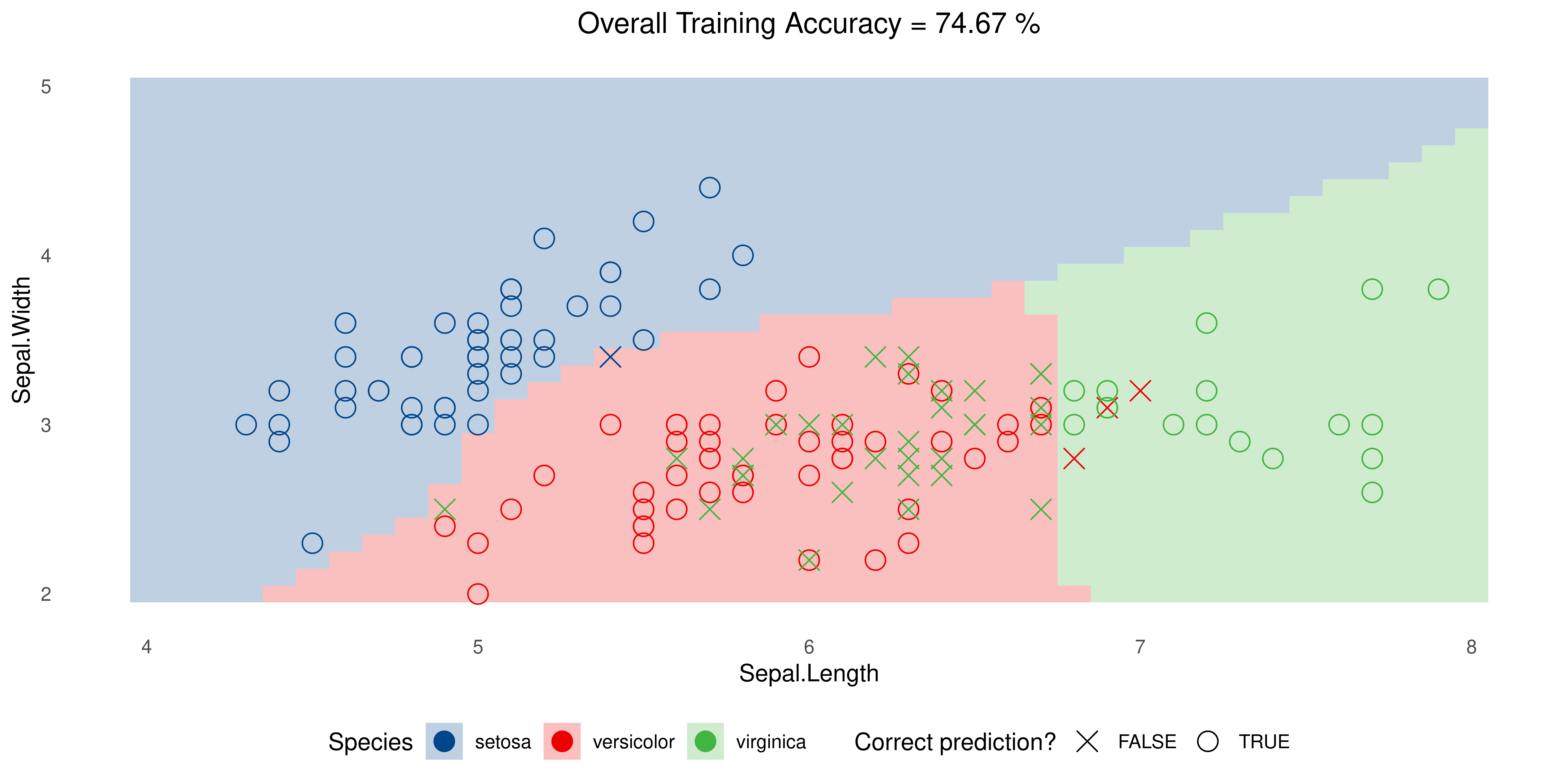
SVM with Sigmoid Kernel
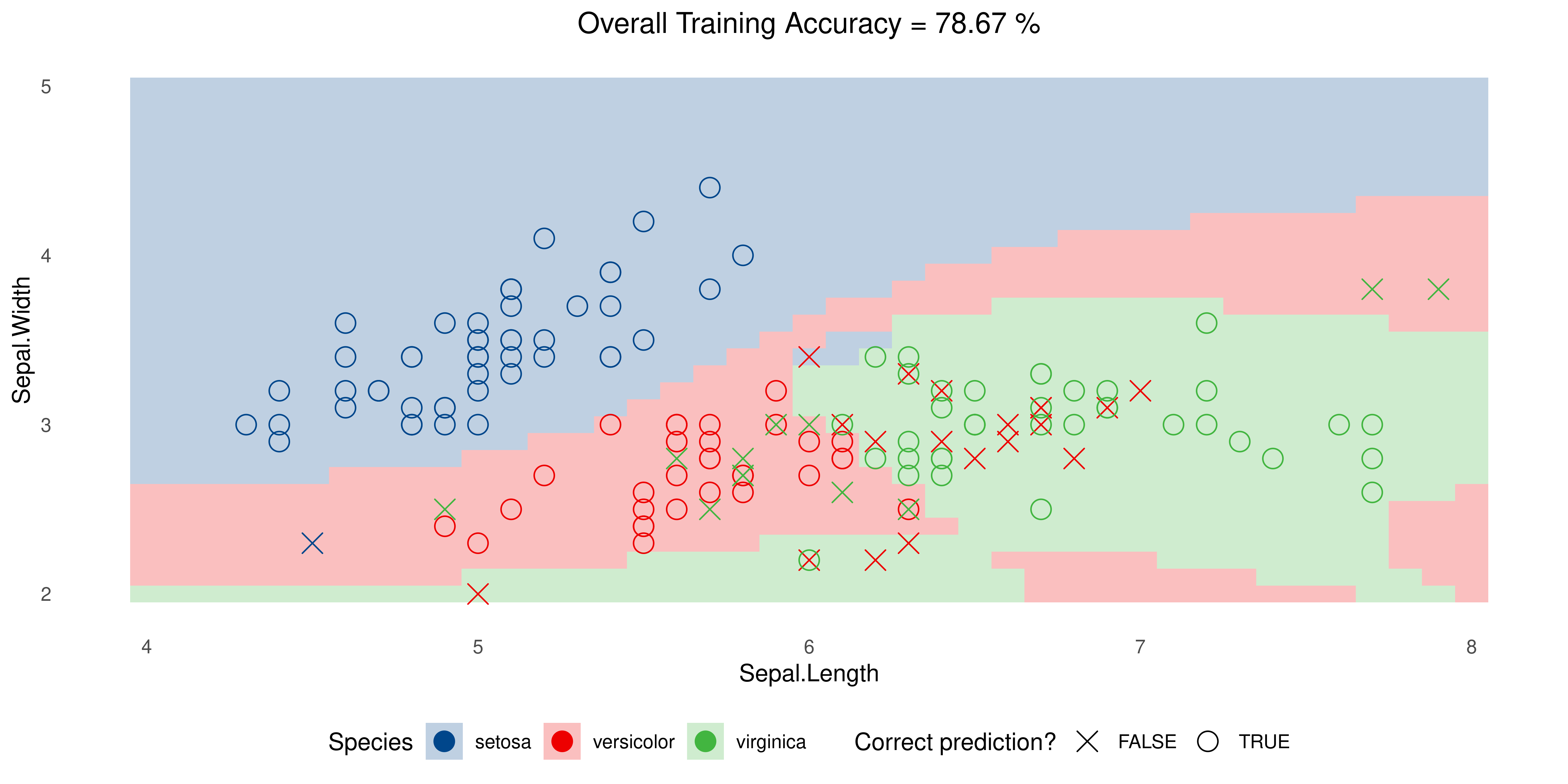
SVM with Radial Kernel
Source Code
Tip
- Use the code below to replicate the plot from the previous slide.
- Found a bug? Report it on Slack.
library(datasets) # to load the iris data
library(tidyverse) # to use things like the pipe (%>%), mutate and if_else
library(ggsci) # just for pretty colours! It enables functions scale_fill_lancet() and scale_colour_lancet().
library(e1071) # to load the SVM algorithm
data(iris) # load the dataset `iris`
# Train the model! change the parameter `kernel`. It accepts 'linear', 'polynomial', 'radial' and 'sigmoid'
model <- svm(Species ~ Sepal.Length + Sepal.Width, data = iris, kernel = 'linear')
# Generate all possible combinations of Sepal.Length and Sepal.Width
kernel.points <- crossing(Sepal.Length = seq(4, 8, 0.1), Sepal.Width = seq(2, 5, 0.1)) %>% mutate(pred = predict(model, .))
# Create a dataframe just for plotting (with predictions)
plot_df <- iris %>% mutate(pred=predict(model, iris), correct = if_else(pred == Species, TRUE, FALSE))
plot_df %>%
ggplot() +
geom_tile(data = kernel.points, aes(x=Sepal.Length, y=Sepal.Width, fill = pred), alpha = 0.25) +
geom_point(aes(x=Sepal.Length, y=Sepal.Width, colour = Species, shape = correct), size = 4) +
scale_shape_manual(values = c(4, 1)) +
scale_colour_lancet() +
scale_fill_lancet() +
theme_minimal() +
theme(panel.grid = element_blank(), legend.position = 'bottom', plot.title = element_text(hjust = 0.5)) +
labs(x = 'Sepal.Length', y = 'Sepal.Width', fill = 'Species', colour = 'Species', shape = 'Correct prediction?',
title = sprintf('Overall Training Accuracy = %.2f %%', 100*(sum(plot_df$correct)/nrow(plot_df))))Overfitting
Overfitting illustrated
Simple models can sometimes be better than complex models.
Simple model on training set
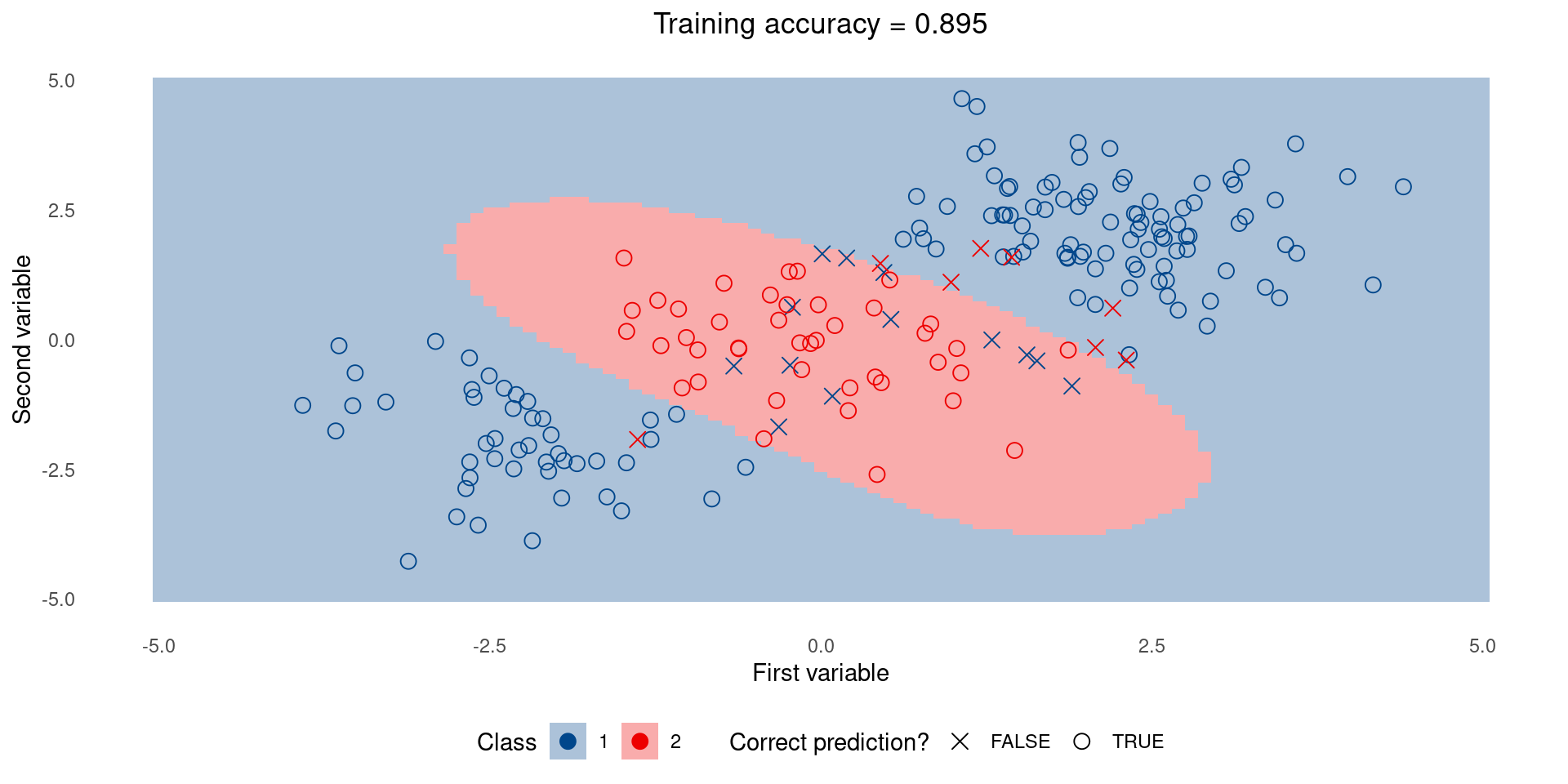
Complex model on training set
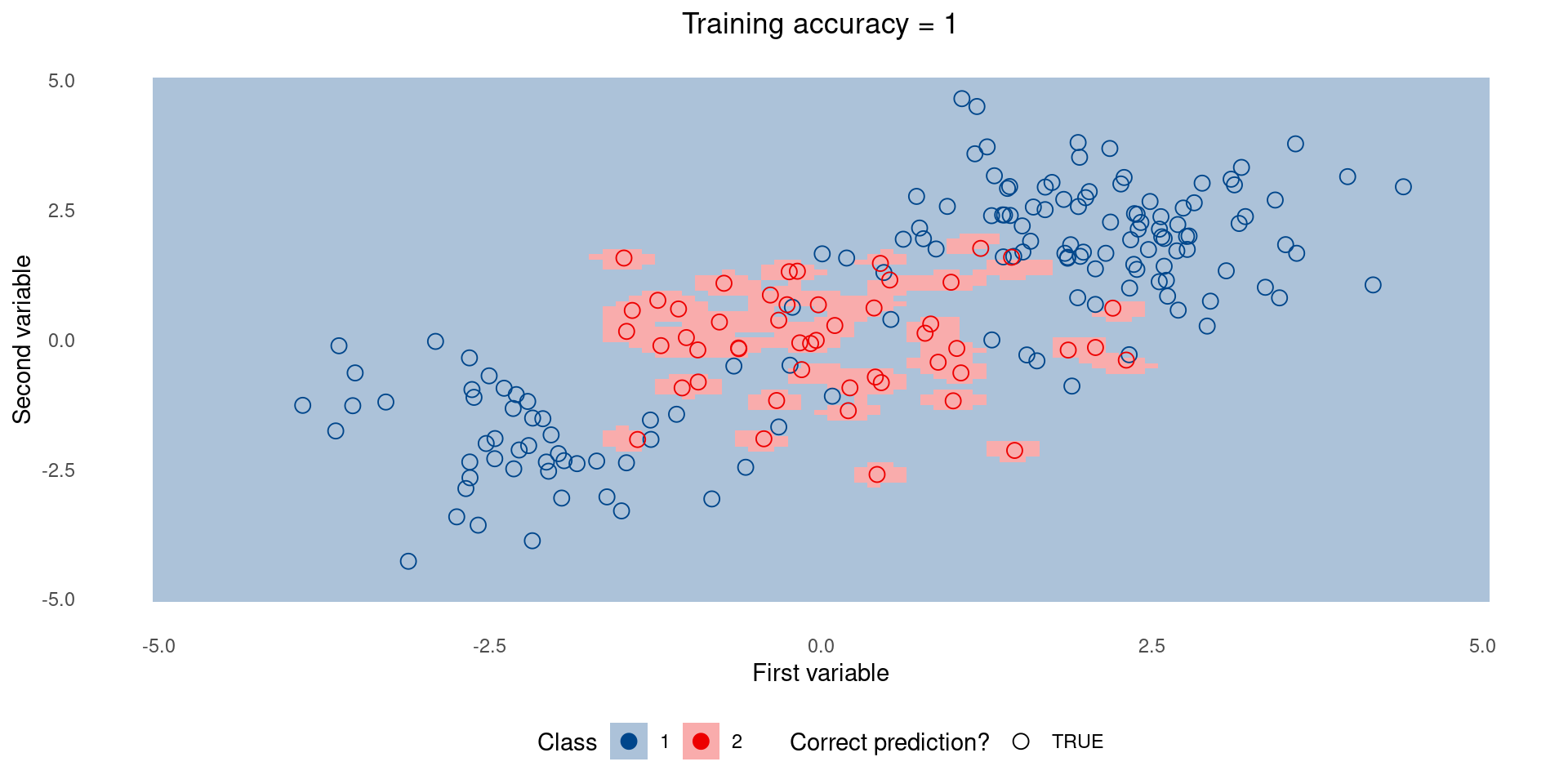
Now let’s create some testing data
Look what happens when I set a different seed (nothing else changes) to construct a test set.
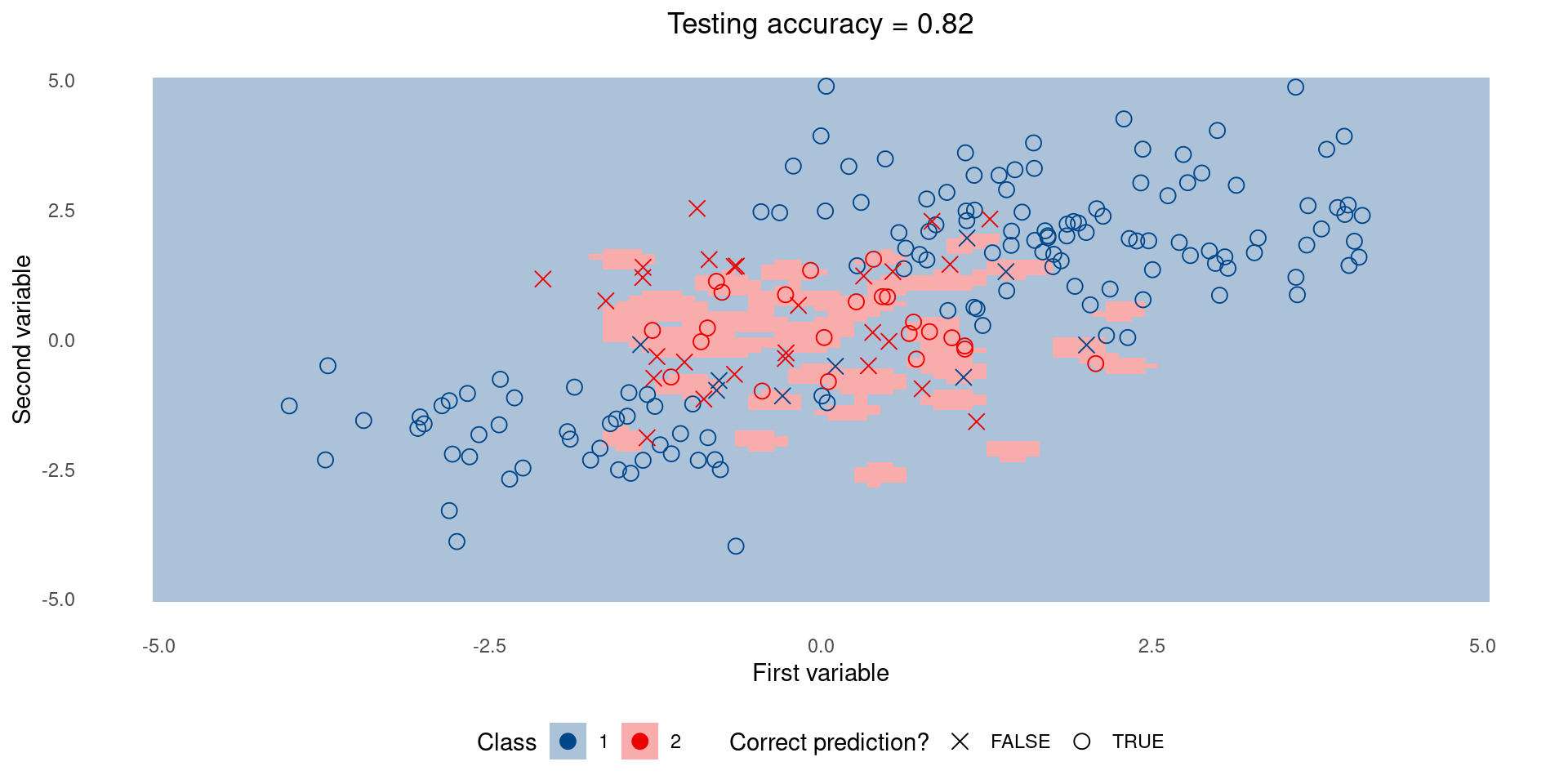
Simple model on test set

Complex model on test set
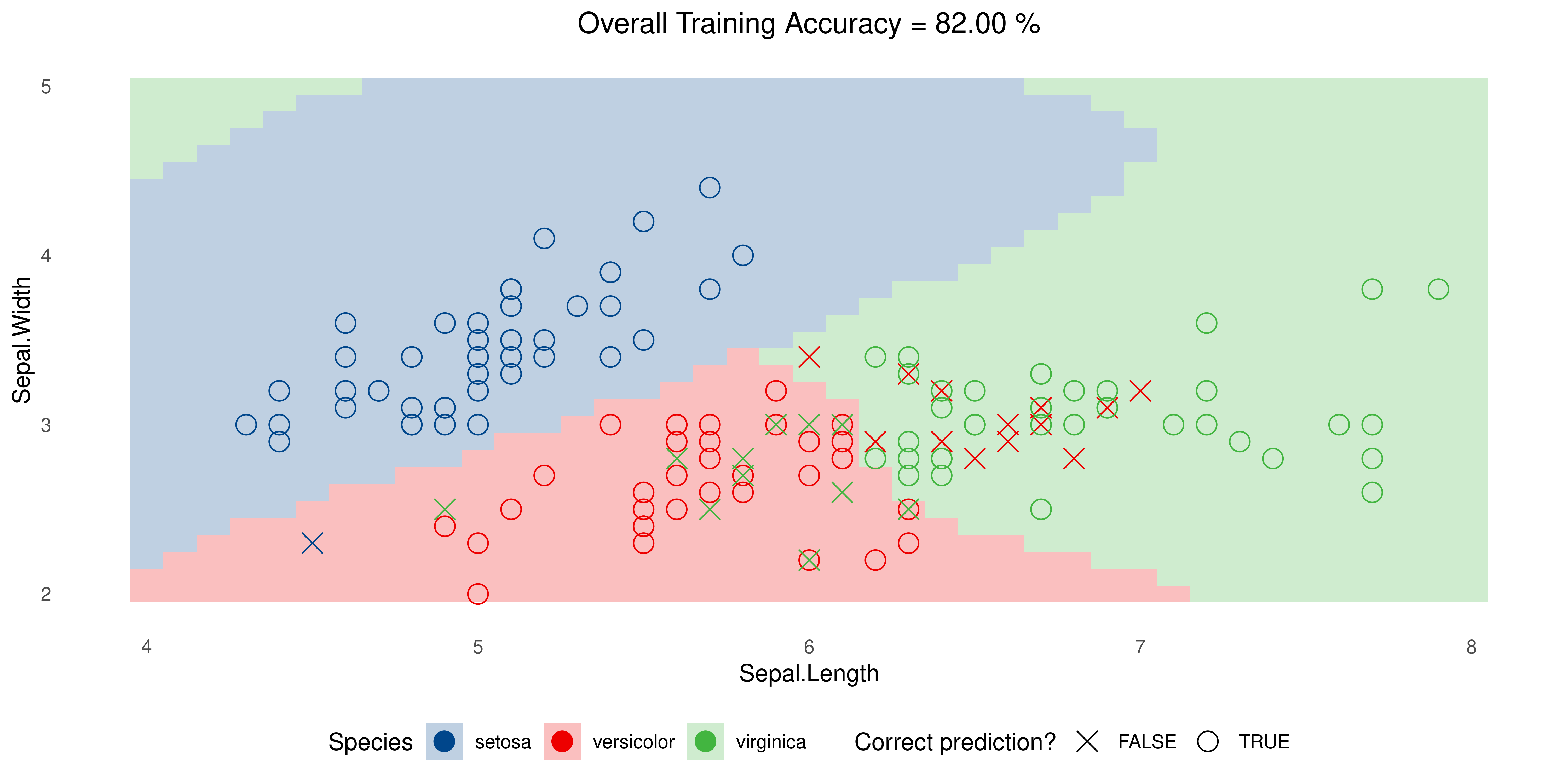
Back to iris
SVM with Radial Kernel but tweaking parameters, namely cost and gamma:
Source Code
Tip
- The only difference is that we added
gammaandcost:
library(datasets) # to load the iris data
library(tidyverse) # to use things like the pipe (%>%), mutate and if_else
library(ggsci) # just for pretty colours! It enables functions scale_fill_lancet() and scale_colour_lancet().
library(e1071) # to load the SVM algorithm
data(iris) # load the dataset `iris`
# Train the model! change the parameter `kernel`. It accepts 'linear', 'polynomial', 'radial' and 'sigmoid'
model <- svm(Species ~ Sepal.Length + Sepal.Width, data = iris, kernel = 'radial', gamma = 10^2, cost = 10^4)
# Generate all possible combinations of Sepal.Length and Sepal.Width
kernel.points <- crossing(Sepal.Length = seq(4, 8, 0.1), Sepal.Width = seq(2, 5, 0.1)) %>% mutate(pred = predict(model, .))
# Create a dataframe just for plotting (with predictions)
plot_df <- iris %>% mutate(pred=predict(model, iris), correct = if_else(pred == Species, TRUE, FALSE))
plot_df %>%
ggplot() +
geom_tile(data = kernel.points, aes(x=Sepal.Length, y=Sepal.Width, fill = pred), alpha = 0.25) +
geom_point(aes(x=Sepal.Length, y=Sepal.Width, colour = Species, shape = correct), size = 4) +
scale_shape_manual(values = c(4, 1)) +
scale_colour_lancet() +
scale_fill_lancet() +
theme_minimal() +
theme(panel.grid = element_blank(), legend.position = 'bottom', plot.title = element_text(hjust = 0.5)) +
labs(x = 'Sepal.Length', y = 'Sepal.Width', fill = 'Species', colour = 'Species', shape = 'Correct prediction?',
title = sprintf('Overall Training Accuracy = %.2f %%', 100*(sum(plot_df$correct)/nrow(plot_df))))Cross-validation* for the rescue!
Example
- A 5-fold cross-validation:
Split 1:
Test
Train
Train
Train
Train
Split 2:
Train
Test
Train
Train
Train
Split 3:
Train
Train
Test
Train
Train
Split 4:
Train
Train
Train
Test
Train
Split 5:
Train
Train
Train
Train
Test
We experimented with k-fold CV in 🗓️ Week 04’s lecture/workshop
Recommendations
- Readings - Decision Trees: (James et al. 2021, sec. 8.1) (ignore section 8.2) - SVMs: (James et al. 2021, chap. 9) - Read it all once just to retain main concepts - Focus your attention on the concepts on the margins of the book
- Practice the code that is contained in these slides! Several times! - There are many datasets in the
ISLR2package (link) - Load any dataset and explore it with these new algorithms
Tips for Summative Problem Set 01
Thank you!

DS202 - Data Science for Social Scientists 🤖 🤹